deepTS
Launch Galaxy| Public Server: | deepTS Server (CN) |
|---|---|
| Container: | deepTS Docker |
| Scope: | Tool Publishing |
| Summary: | Explore transcriptional switches from pairwise, temporal, and population RNA-Seq using deepTS. |
Comments
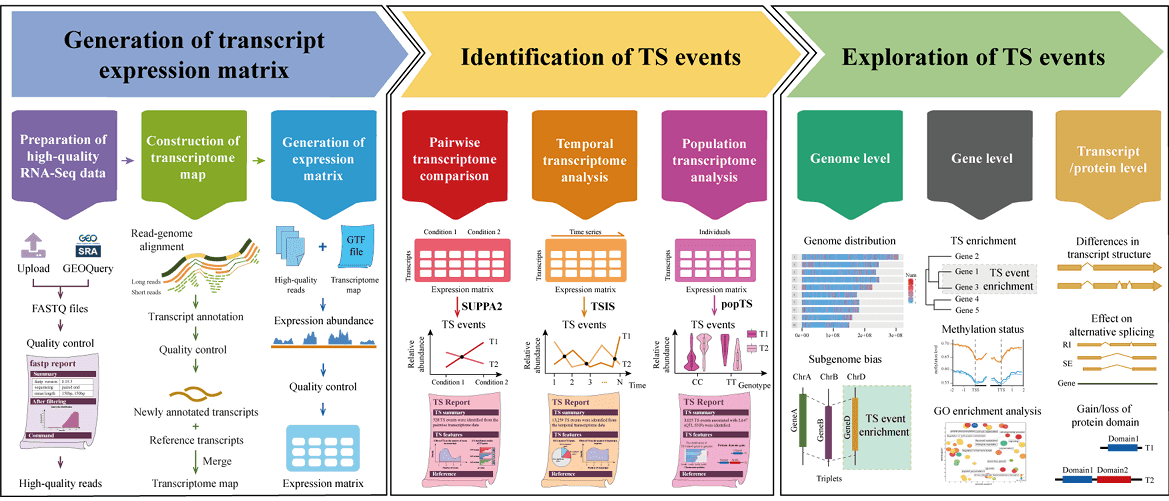
- deepTS is a powerful and flexible web-based Galaxy platform for identifying, visualizing and analyzing transcriptional switch (TS) events from pairwise, temporal and population transcriptome data. deepTS consists of three main functional modules, covering the processes of read cleaning and mapping, transcriptome map construction, expression abundance estimation, multiple-condition TS analysis pipelines, multiple-level TS characterization, and multiple-form visualization to allow users to perform TS analysis using either raw RNA-Seq data or expression abundance matrix directly.
User Support
- For comments/suggestions/error reports, please contact Siyuan Chen (chenzhuod@gmail.com) or or Zhixu Qiu (zhixuqiu2015@gmail.com).
- Docker Installation Insstructions
- Tutorial
- Test datasets are available
Citations
- Qiu, Z., Chen, S., Qi, Y., Liu, C., Zhai, J., Xie, S., & Ma, C. (2020). Exploring transcriptional switches from pairwise, temporal and population RNA-Seq data using deepTS. Briefings in Bioinformatics. doi: 10.1093/bib/bbaa137
- deepTS tagged publications in the Galaxy Publication Library
Sponsors
- deepTS is developed and maintained by the lab of Prof. Chuang Ma at the Center of Bioinformatics, College of Life Sciences, Northwest A&F University.