
Welcome to the microGalaxy Community
Your hub for microbiological data analysis in Galaxy, whether you're analyzing microbiome samples, bacterial isolates, long or short reads, shotgun or 16S data, genomics, transcriptomics, proteomics, metabolomics, or multi-omics integrative analyses.
Community Goals
Since 2021, the microGalaxy community has focused on:
- Coordinating efforts across the Galaxy ecosystem to minimize redundancy in tools, workflows, and training.
- Sharing advancements within and beyond the Galaxy community.
- Supporting collaboration among users, developers, and trainers.
We also aim to:
- Develop and maintain microbiological data analysis tools and workflows in Galaxy.
- Standardize best practices for FAIR (Findable, Accessible, Interoperable, Reusable) microbiological data analysis.
- Expand documentation and training to make learning accessible for all skill levels.
Who Are We?
We are a diverse group of users, developers, trainers, and bioinformaticians passionate about advancing microbiological data analysis in Galaxy. Everyone is welcome!
- Developers: Developing microbiological tools or workflows in Galaxy? Looking for collaborators or testers? Join us!
- Users: Analyzing microbiome samples, bacterial isolates, or multi-omics data? Share your feedback, test new resources, or request tools.
- Trainers: Using Galaxy for training? Help us improve resources and support users.
- Bioinformaticians: Contribute your expertise to ensure our tools reflect the latest best practices.
Community Coordinators:
- Bérénice Batut (Institut Français de Bioinformatique & AuBi, France)
- Paul Zierep (University of Freiburg, Germany)
Getting Started
The Microbiology Galaxy Lab (MGL)
The MGL is a specialized Galaxy extension designed for microbiology research. It features:
- A curated tool panel with community-approved tools organized by analysis type.
- A central resource hub with direct links to tools, workflows, and training materials for microbial isolates and microbiome data.
- Community-developed and FAIR-compliantworkflows.
- Quick access to support and the microGalaxy community.
Ready to explore? Log in with your Galaxy account on the server closest to you:
- United States:microbiology.usegalaxy.org
- Europe:microbiology.usegalaxy.eu
- Australia:microbiology.usegalaxy.org.au
- France:microbiology.usegalaxy.fr
Training
The Galaxy Training Network (GTN) offers 40+ tutorials, 15 videos (16 hours), and 3 structured learning pathways for microbiology analyses—from basic sequence analysis to advanced metaproteomics and metagenomics.
Community Maintained, Curated and Ready-to-Use Resources
Tools and Workflows
MGL provides 315+ tool suites and 115+ curated workflows for comprehensive microbial data analysis:
 Comprehensive workflows for microbial isolate and microbiome analysis.
Comprehensive workflows for microbial isolate and microbiome analysis.
Tools
Explore the 300+ tool suites (880+ individual tools) for microbiological data analysis available in the ToolShed and curated by the community:
This list is powered by the Galaxy Codex Project and regularly updated and curated by the microGalaxy community to provide state-of-the-art tools. You can explore the raw metadata.
Highlights:
- Integration of complete frameworks (e.g.QIIME 2, MetaPhlAn, and soon Anvi'o) for intuitive access to extensive resources.
- 170+ reference genomes and 11TB of reference data for taxonomic classification and functional annotation.
- Interactive tools like Pavian, Phinch, Shiny Phyloseq, Apollo, Jupyter Notebooks, and RStudio.
Are we missing a tool, or should some tools be removed? Reach out to us or contribute by modifying the To keep and/or Deprecated column in the curation file to help us keep this list comprehensive.
Workflows
Access to 115+ ready-to-use workflows, e.g. the integration of the MGnify amplicon pipeline:
This list is powered by the Galaxy Codex Project and regularly updated and curated by the microGalaxy community to provide state-of-the-art workflows. You can explore the raw metadata.
Are we missing a public workflow, or should some be removed? Reach out to us or contribute by modifying the To keep and/or Deprecated column in the curation file to help us keep this list comprehensive.
Training Events
The microGalaxy community has organized 30+ training events over the past five years, including the Galaxy Training Academy (formerly Galaxy Smörgåsbord):
Specialized trainings, such as Mycobacterium tuberculosis genomic analysis, have reached global participants, especially in the Global South.
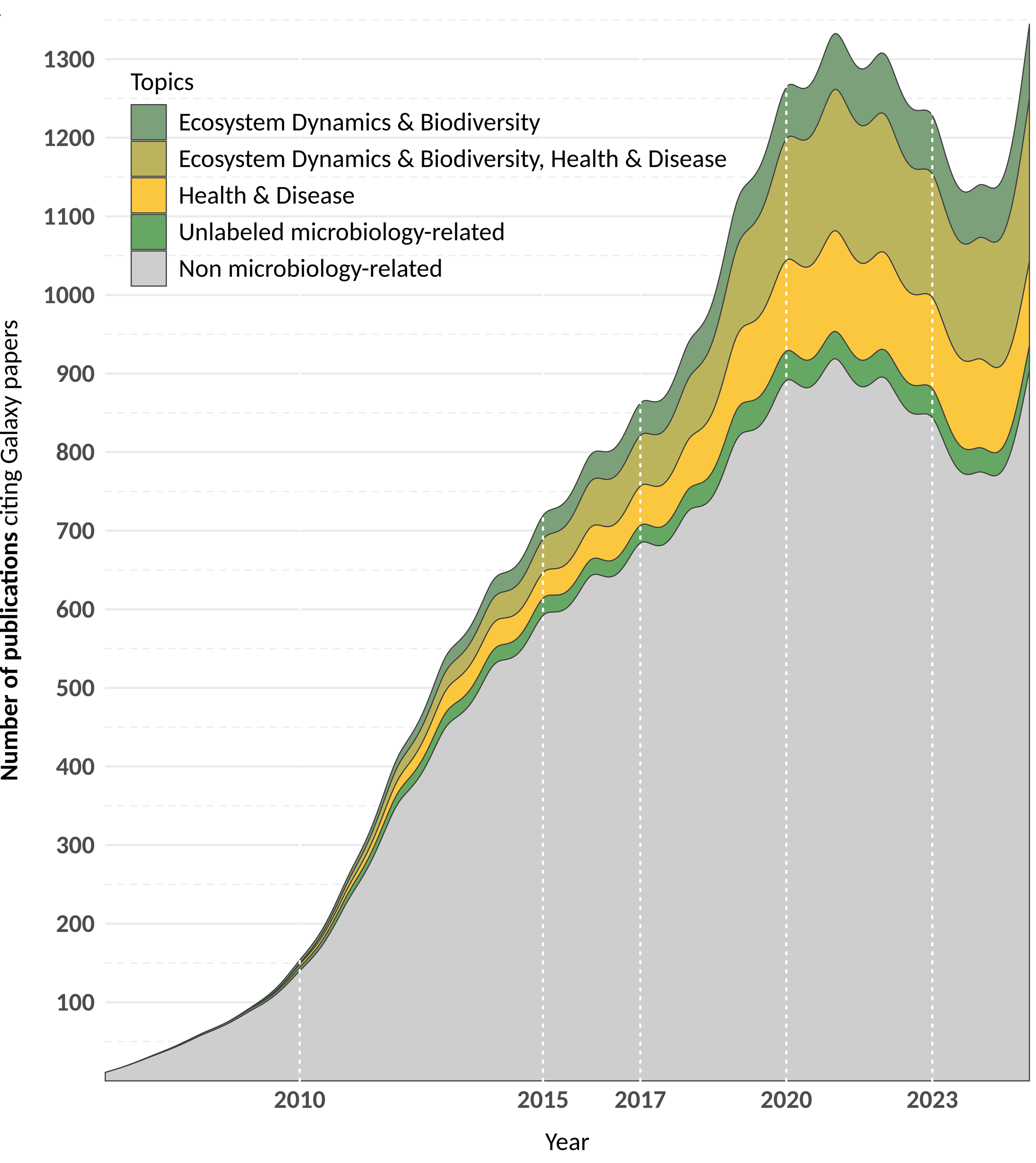
Galaxy Adoption for Microbial Research
Galaxy is a trusted platform for microbial data analysis, with 10,459+ publications using Galaxy, including 2,908 microbiology-focused studies:
- 836 studies (28.75%) on health and disease.
- 593 studies (20.39%) on ecosystems and biodiversity.
- 1,116 studies (38.38%) bridging both domains.
(Meta)genomics is the most common application, followed by metabarcoding and metatranscriptomics. Key analyses include taxonomic classification, functional annotation, and AMR gene profiling.
Applications in Health, Disease, and Ecosystem Research
The microGalaxy community and the Microbiology Galaxy Lab (MGL) resources have been used for diverse research applications in health, disease, and ecosystem studies
Health and Disease
- Pathogen Analysis:, e.g. microbial pathogenesis, host-microbe interactions, and antibiotic resistance mechanisms.
- AMR Research: via platforms like ABRomics and BenchAMRking, which provide workflows for AMR gene identification and clinical reporting.
- Pathogen Detection:, e.g. PathoGFAIR supports outbreak tracking and food safety.
- Clinical Metaproteomics: to analyze microbial proteins in diseases like COVID-19, cystic fibrosis, and ovarian cancer.
- AI and Machine Learning: to discover microbial biomarkers and novel viruses with tools like MetaSBT.
Ecosystem Dynamics and Biodiversity
- Community Dynamic using metabarcoding, metagenomics, and whole-genome sequencing for accurate classification, and explore root microbiomes, beer microorganisms (BeerDEcoded), soil community (BioDIGS), and global mycorrhizal fungi (SPUN).
- Functional Annotation with gene prediction and functional profiling.
- Multi-Omics Integration to link genetic potential with real-time activity, e.g. in studies to study microbe communities in atmospher (Péguilhan et al., 2025) and biogas reactor (Schiml et al., 2023).
Get Involved
The microGalaxy community is open to all! No formal membership required.
Communication Channels
Meetings and Events
- Online Meetings: 4 times/year (alternating times for global participation).
- In-Person Events: Annual Galaxy Community Conference and European Galaxy Days.
- Hackathons and Workshops: Collaborative events to develop workflows, tools and other functionalities, as a community, with other Galaxy communities and working (e.g. with the Intergalactic Workflow Commission (IWC) in 2025), or during external events like the ELIXIR BioHackathon or
Community Calendar
Join Us
- Subscribe to the mailing list.
- Say hello in the Matrix channel.
- Attend a meeting.
Acknowledgments
We thank the microGalaxy community and the global Galaxy community of users, developers, educators, and administrators.
Partner Projects
| Partners | Description | People Involved |
|---|---|---|
| Seq4AMR | Network for Integrating Microbial Sequencing and AMR Platforms | Saskia Hilteman |
| ELIXIR Microbiome Community | Bérénice Batut, Paul Zierep | |
| Galaxy-P | Pratik Jagtap | |
| IRIDA | Integrated Rapid Infectious Disease Analysis | Aaron Petkau |
| ABRomics | French National Multi-Omics Platform for AMR Research | Bérénice Batut, Cléa Siguret |
| FAIRyMAGs | FAIRyMAGs: Optimising Metagenomics Assembled Genomes building | Paul Zierep, Mina Hojat Ansari, Patrick Bühler, Santino Faack, Giuseppe Defazio, Bruno Fosso, Martin Beracochea, Santiago Sanchez, Alexandra Hottmann, Bérénice Batut |