Galaxy Erasmus MC
The homepage of the Erasmus MC Galaxy community
Galaxy is an open-source platform for FAIR data analysis that enables users to:
- use tools from various domains (that can be plugged into workflows) through its graphical web interface.
- run code in interactive environments (RStudio, Jupyter...) along with other tools or workflows.
- manage data by sharing and publishing results, workflows, and visualizations.
- ensure reproducibility by capturing the necessary information to repeat and understand data analyses.
The Galaxy Community is actively involved in helping the ecosystem improve and sharing scientific discoveries.
News
Galaxy and Bioconductor Community Conference Meeting Report
From groundbreaking research to hands-on workshops and live demos, GBCC2025 showcased the best of the Galaxy and Bioconductor communities.
Freiburg Galaxy Team’s Internal Tool Development Workshop
Know more about the recent Tool Development workshop organized at Freiburg Galaxy Team
Scaling Up Hands-On Bioinformatics Training with TIAAS – An Open University Perspective
Back in September 2024, we ran the Open University Bioinformatics Bootcamp—a free, five-day online course introducing students to the core tools and techniques used in single-cell biology
SURF Research Cloud as infrastructure for Galaxy in The Netherlands
Dutch scientists can now use Galaxy in secure environments.
Enhancing Scientific Training: The Galaxy Training Network’s Role in the ELIXIR Training Life-Cycle
In the rapidly evolving landscape of data science, continuous learning and skill development are crucial
Events
Aug 5BRC Measles Bioinformatic Analysis Webinar
Please join the NCBI and the NIAID-funded BRCs to discuss Measles data analysis!
Aug 19 - Aug 212025 NCI Informatics Technology for Cancer Research Annual Meeting
Join the 2025 NCI ITCR Annual Meeting to explore cutting-edge cancer informatics tools and collaborations.
Aug 26 - Aug 28Galaxy @ CoRDI 2025
Do you want to learn how Galaxy can help you with your research and reproducible research data management? Join us at CoRDI!
FTP Upload
For large files, FTP upload is also available on this server. Connect to port 23 with the same credentials you use to log into Galaxy. Once uploaded, open the Galaxy file upload menu, and click on the Choose FTP file button to import the files into your history.
As an example, a screenshot of the configuration settings for FileZilla can be found here.
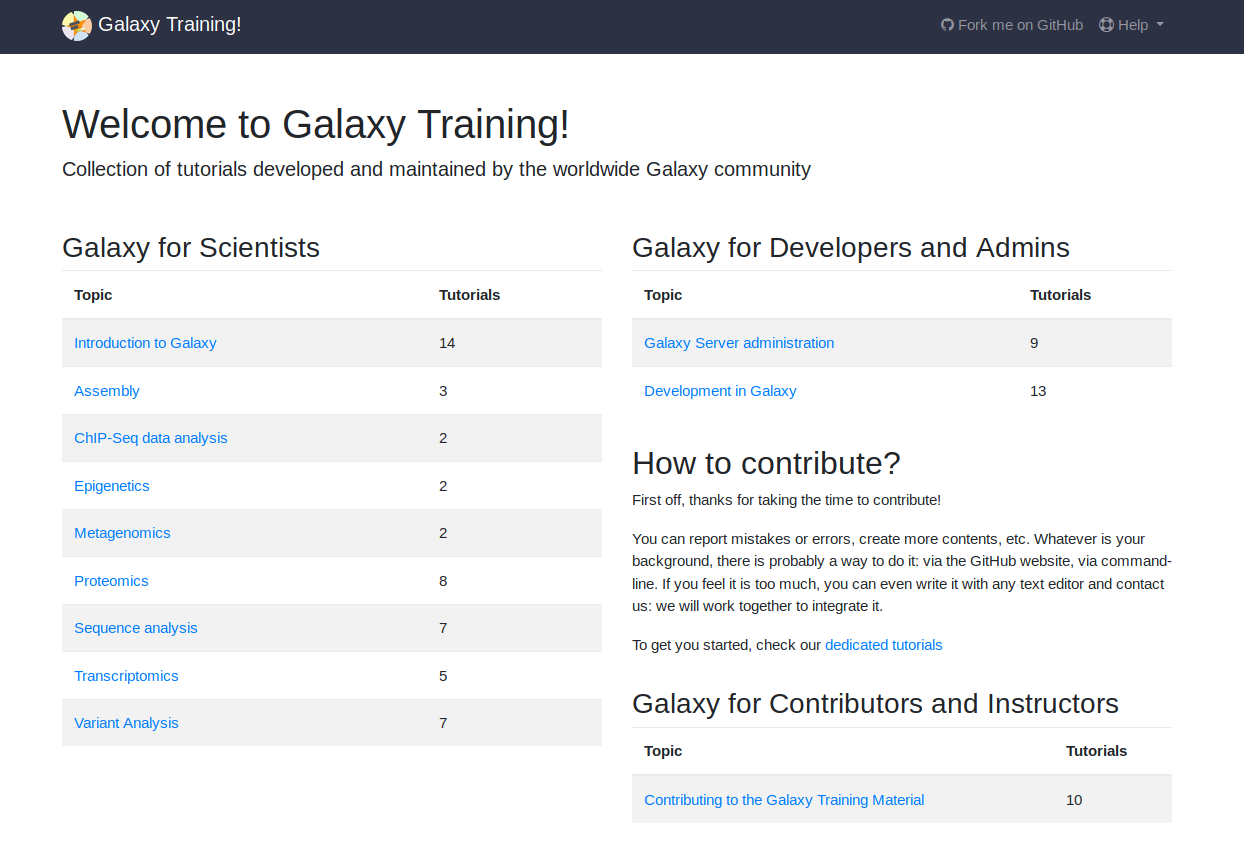
Training
We regularly provide workshops.
But we cannot always meet capacity, so we've put all of our training materials online. This has become a community project with people from all over the world contributing training materials.
Topics include: variant analysis, transcriptomics, metagenomics, epigenetics, and many more!
Our team
This Galaxy is maintained by the Bioinformatics group of the department of Pathology.
For any questions regarding this Galaxy server, please contact us.