Bioconda: sustainable and comprehensive software distribution for the life sciences
3 years ago the Galaxy community evaluated mechanism to make software installation more robust and we joined the Bioconda community, to fix the software deployment problem once and for all. Today, we are happy to annouce that the Bioconda manuscript is now published in the latest Nature Methods issue (public view). usegalaxy.eu and many other Galaxy instance are already powered by Bioconda and you might use it daily without knowing :)
Abstract
Bioinformatics software comes in a variety of programming languages and requires diverse installation methods. This heterogeneity makes management of a software stack complicated, error-prone, and inordinately time-consuming. Whereas software deployment has traditionally been handled by administrators, ensuring the reproducibility of data analyses1,2,3 requires that the researcher be able to maintain full control of the software environment, rapidly modify it without administrative privileges, and reproduce the same software stack on different machines.
The Conda package manager (https://conda.io) has become an increasingly popular means to overcome these challenges for all major operating systems. Conda normalizes software installations across language ecosystems by describing each software with a human readable ‘recipe’ that defines meta-information and dependencies, as well as a simple ‘build script’ that performs the steps necessary to build and install the software. Conda builds software packages in an isolated environment, transforming them into relocatable binaries. Importantly, it obviates reliance on system-wide administration privileges by allowing users to generate isolated software environments in which they can manage software versions by project, without generating incompatibilities and side-effects (Supplementary Results). These environments support reproducibility, as they can be rapidly exchanged via files that describe their installation state. Conda is tightly integrated into popular solutions for reproducible data analysis such as Galaxy, bcbio-nextgen (https://github.com/chapmanb/bcbio-nextgen), and Snakemake. To further enhance reproducibility guarantees, Conda can be combined with container or virtual machine-based approaches and archive facilities such as Zenodo (Supplementary Results). Finally, although Conda provides many commonly used packages by default, it also allows users to optionally include additional, community-managed repositories of packages (termed channels).
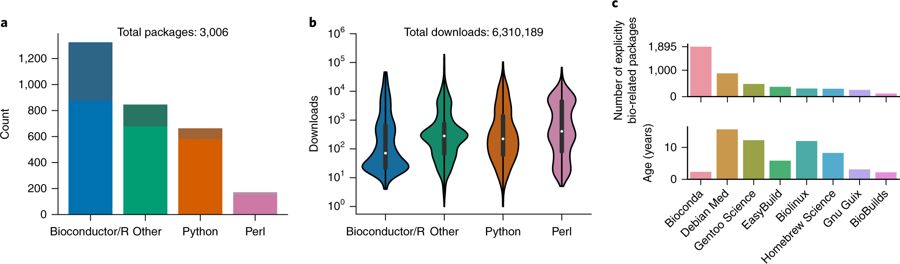
To unlock the benefits of Conda for the life sciences, we present the Bioconda project (https://bioconda.github.io). The Bioconda project provides over 3,000 (Updated: 4000!!!) Conda software packages for Linux and macOS. Rapid turnaround times (Supplementary Results) and extensive documentation (https://bioconda.github.io/contributing.html) have led to a growing community of over 200 international scientists working in the project (Supplementary Results). The project is led by a core team, which is complemented by interest groups for particular language ecosystems. Unlimited (in time and space) storage for generated packages is donated by Anaconda Inc. All other used infrastructure is free of charge. Bioconda provides packages from various language ecosystems such as Python, R (CRAN and Bioconductor), Perl, Haskell, Java, and C/C++ (Fig. 1a). Many of the packages have complex dependency structures that require various manual steps for installation when not relying on a package manager like Conda (Supplementary Results). With over 6.3 million downloads, Bioconda has become a backbone of bioinformatics infrastructure that is used heavily across all language ecosystems (Fig. 1b). It is complemented by the conda-forge project (https://conda-forge.github.io), which hosts software not specifically related to the biological sciences. This separation has proven beneficial, because the focused nature of the Bioconda community allows for fast turnaround times and support when a user needs to contribute packages or fix problems. Nevertheless, the two projects collaborate closely, and the Bioconda team maintains over 500 packages hosted by conda-forge.

