The Galaxy Genome Annotation Project - Scaling Genome Annotation for the Masses
Scaling Genome Annotation for the Masses
The Galaxy Genome Annotation (GGA) Project is focused on supporting genome annotation inside Galaxy. It consists of several teams, projects, and tool suites that are working closely together to deliver a comprehensive, scalable and easy to use Genome Annotation experience. This blog post will highlight a few of the recent developments and the new Genome Annotation subsection of the European Galaxy server: https://annotation.usegalaxy.eu

This project was recently presented by Helena Rasche at ISMB/ECCB 2019.
Collaborative Annotation
Apollo is a web service for Collaborative Annotation - a “Google Docs of Genome Annotation” if you like to think in Google Terms. It allows for real-time, collaborative genome annotation, editing, and review of your favorite genome. Genome Annotation requires manual curation and review many hands are needed - Biology is a team sport! And Apollo is a wonderful tool to support your team.
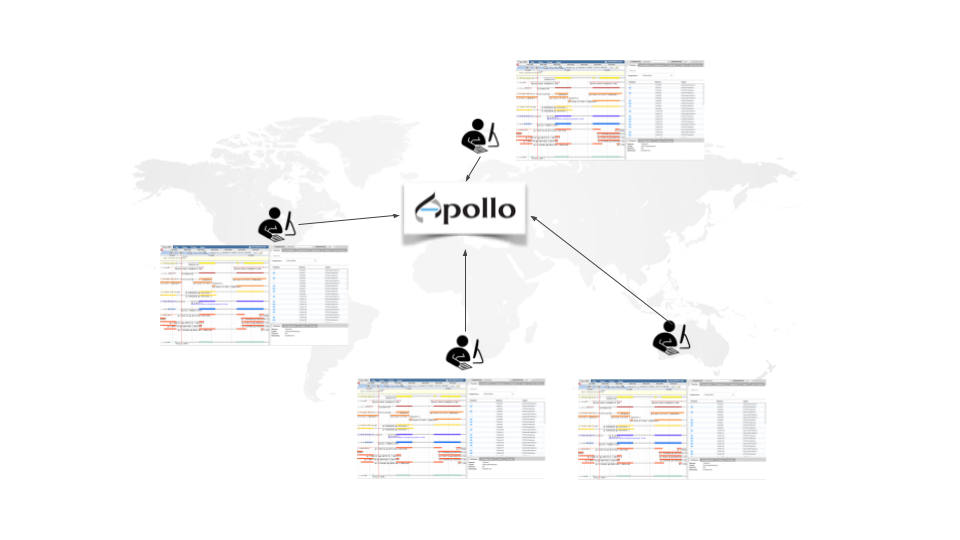
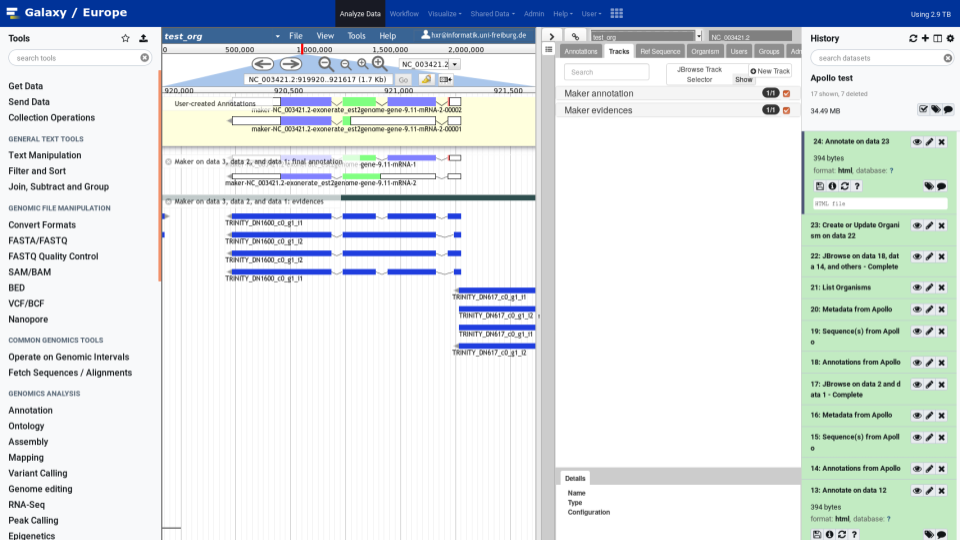
To make your life easy we worked closely with the Apollo team and bridged Galaxy with Apollo. You can do automatic annotation with Galaxy, using its powerful workflow system, and afterwards send the data to Apollo for manual curation and collaborative editing! A typical Galaxy-Apollo use-case might look like that:
- Fetch Data
- Analyse in Galaxy
- Send to Apollo
- Collaboratively Annotate → repeat
Tools
The European Galaxy server for Genome Annotation contains a wide array of high-profile tools for structural and functional annotation. Below is a (incomplete) list of tools to give you an impression of what is possible currently in Galaxy.
-
Assembly
- Spades
- Mira
-
Structural Prediction
- Glimmer
- Augustus
-
Functional Prediction
- BLAST+
- InterProScan
- BLAST
- Diamond
-
Utilities
- FASTA manipulation
- EMBOSS
-
Comparative Genomics
- CD-Hit, ClustalW
- AntiSmash
- mummer
- Roary
-
Annotation / Viz.
- Apollo Tools
- JBrowse-in-Galaxy
- Circos
-
Automatic annotation
- MAKER
- Prokka
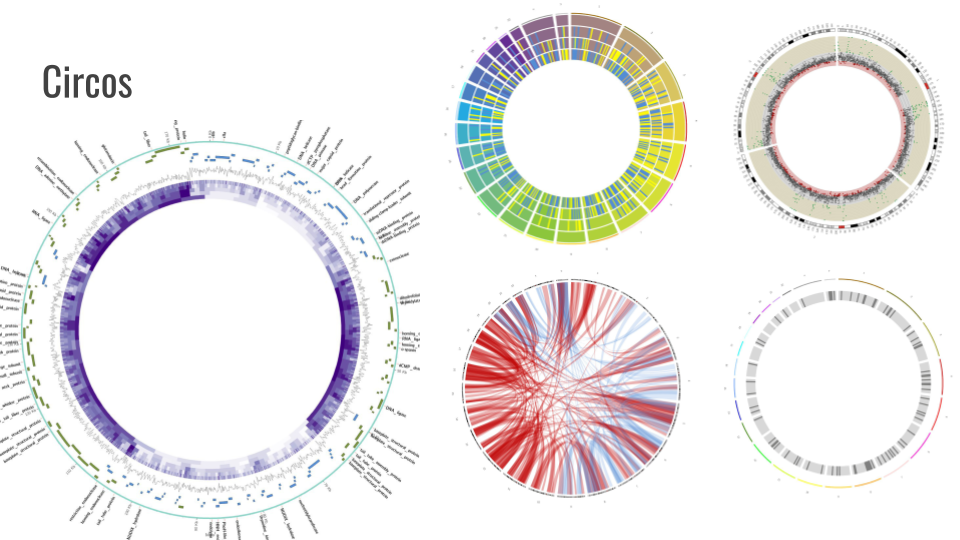
Circos
Circos is a software package for visualizing data and information. It visualizes data in a circular layout — this makes Circos ideal for exploring relationships between objects or positions. There are other reasons why a circular layout is advantageous, not the least being the fact that it is attractive. The Circos tool is in final polishing, we will integrate it into training and annotation workflows over the next two months.
InterProScan
InterProScan is a batch tool to query the Interpro database. It provides annotations based on multiple searches of profile and other functional databases:
- TIGRFAM: protein families based on Hidden Markov Models or HMMs
- PIRSF: non-overlapping clustering of UniProtKB sequences into a hierarchical order (evolutionary relationships)
- ProDom: set of protein domain families generated from the UniProtKB
- Panther: Protein ANalysis THrough Evolutionary Relationships
- SMART: identification and analysis of domain architectures based on Hidden Markov Models or HMMs
- PROSITE Profiles: protein domains, families and functional sites as well as associated profiles to identify them
- PROSITE Pattern: protein domains, families and functional sites as well as associated patterns to identify them
- HAMAP: High-quality Automated Annotation of Microbial Proteomes
- PfamA: protein families, each represented by multiple sequence alignments and hidden Markov models
- PRINTS: group of conserved motifs (fingerprints) used to characterise a protein family
- SUPERFAMILY: database of structural and functional annotation
- Coils: Prediction of Coiled Coil Regions in Proteins
- Gene3d: Structural assignment for whole genes and genomes using the CATH domain structure database
- SignalP Gram Positive Bacteria
- SignalP Gram Negative Bacteria
- SignalP Eukaryotic Bacteria
- Phobius: combined transmembrane topology and signal peptide predictor
- TMHMM: Prediction of transmembrane helices in proteins
MAKER
MAKER is a portable and easily configurable genome annotation pipeline. Its purpose is to allow smaller eukaryotic and prokaryotic genome projects to independently annotate their genomes and to create genome databases. MAKER identifies repeats, aligns ESTs and proteins to a genome, produces ab-initio gene predictions and automatically synthesizes these data into gene annotations having evidence-based quality values. MAKER is also easily trainable: outputs of preliminary runs can be used to automatically retrain its gene prediction algorithm, producing higher quality gene-models on seusequent runs. MAKER's inputs are minimal and its ouputs can be directly loaded into a GMOD database. They can also be viewed in the Apollo genome browser; this feature of MAKER provides an easy means to annotate, view and edit individual contigs and BACs without the overhead of a database. MAKER should prove especially useful for emerging model organism projects with minimal bioinformatics expertise and computer resources. Try MAKER in Galaxy!
Prokka
Prokka is a software tool to rapidly annotate bacterial, archaeal and viral genomes, and produce output files that require only minor tweaking to submit to GenBank/ENA/DDBJ. Try Prokka in Galaxy!
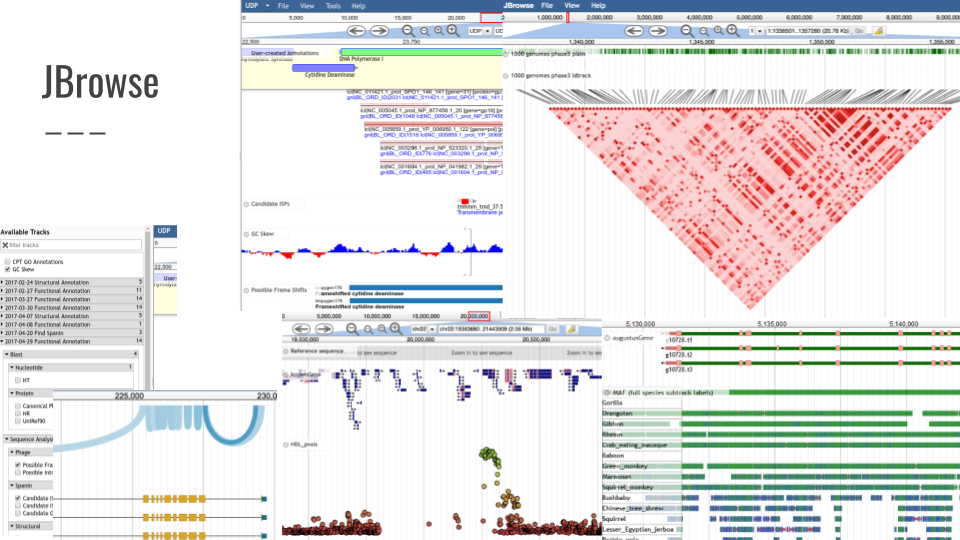
JBrowse
JBrowse is a fast, embeddable genome browser built completely with JavaScript and HTML5.
Functional and Structural Annotation
Functional annotation with a variety of different tools working together and finally handing of the results to Apollo.
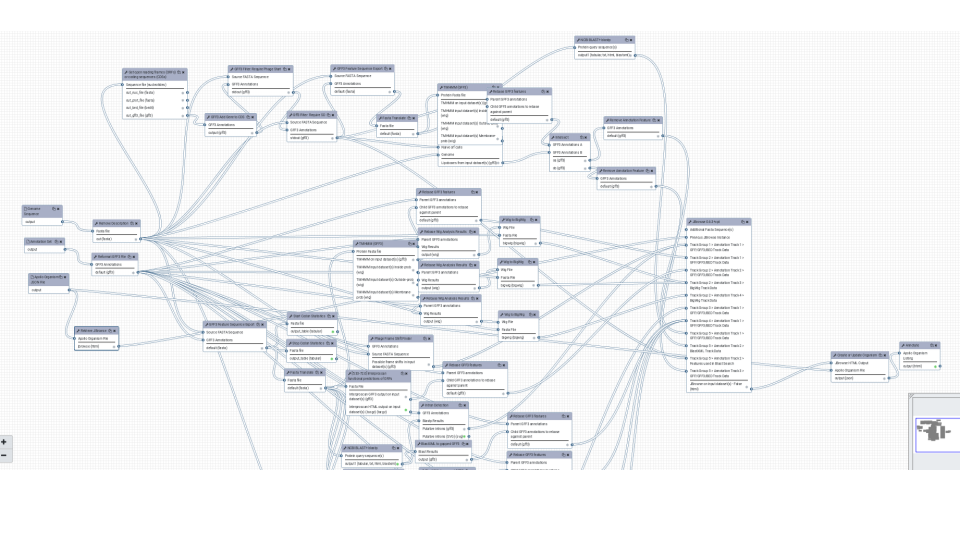
The same for structural annotation of your Genome. The results will be send to Apollo to enable collaborative editing!
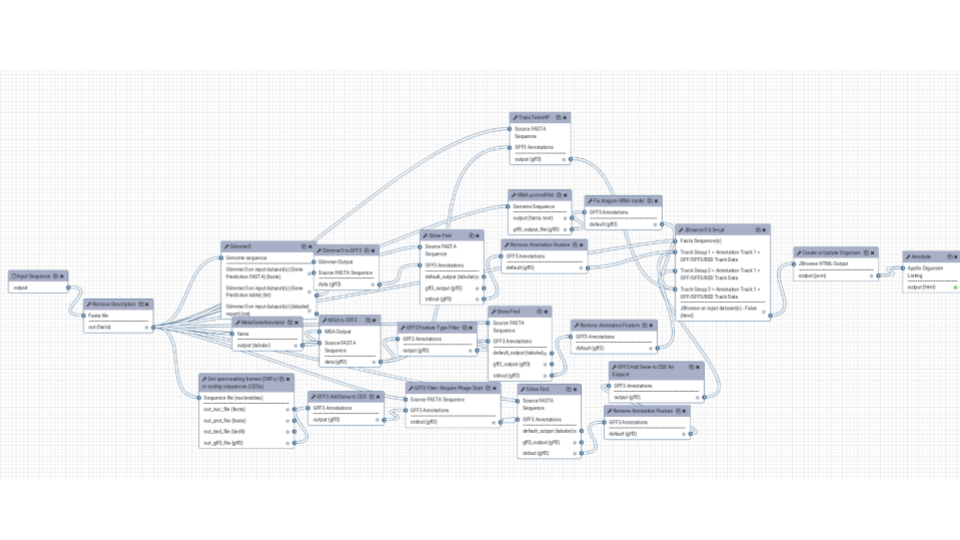
Training
As usual the Galaxy community is providing hands-on training material to get your started quickly. Check out our Genome Annotation trainings at: https://training.galaxyproject.org/training-material/topics/genome-annotation/ and learn more about MAKER, BUSCO, SNAP/Augustus, Apollo, Tripal querying + admin, Chado querying, JBrowse and Circos.
Thanks!
Thanks to Anthony Bretaudeau (BIPAA/GenOuest), Nathan Dunn (Apollo Lead), Helena Rasche (Galaxy Europe) and all the fantastic contributors from all over the world (Ian Holmes, Cory Maughmer, Peter van Heusen, Suzanna Lewis, Eduardo de Paiva Alves, Torsten Seemann, Simon Gladmann, Loraine Brillet-Guéguen, Matéo Boudet, Luke Sargent, Björn Grüning …)