ChiRA tool suite - an integrated framework for Chimeric Read Analysis from RNA-RNA interactome data
an integrated framework for Chimeric Read Analysis from RNA-RNA interactome data
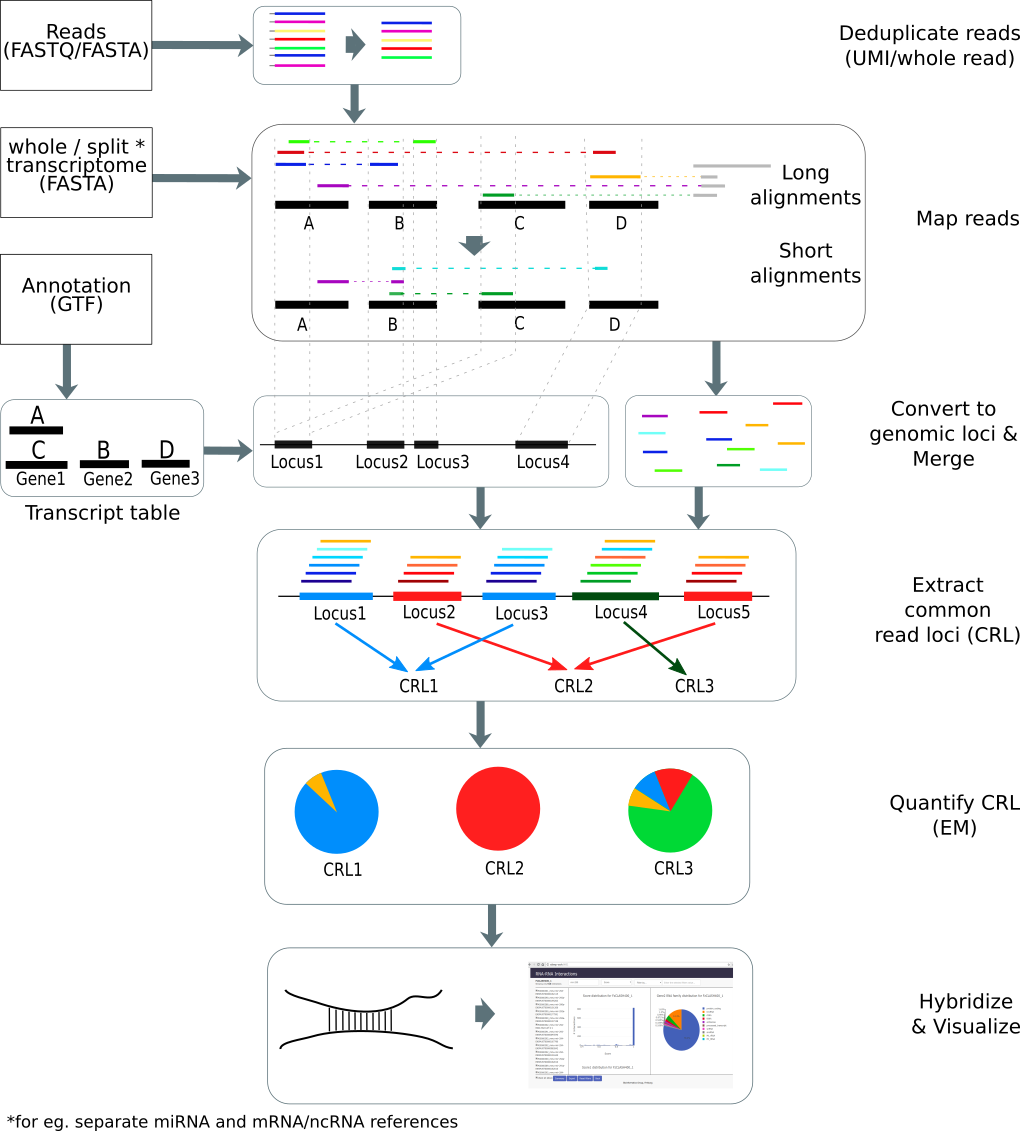
ChiRA is a set of tools to analyze RNA-RNA interactome experimental data such as CLASH, CLEAR-CLIP, PARIS, LIGR-Seq, etc.
The reads from these datasets are chimeric, i.e, two interacting RNA fragments fused into a single read. Limitations of the current library preparation protocols limit the length of each sequenced interacting RNA fragment. These smaller RNA fragments are often harder to map considering that the boundaries of each RNA fragment in the read are unknown. The ChiRA tool suite maps these reads sensitively and infers the true origins of them by quantifying the mapped loci. The visualization framework ChiRAViz gives flexibility in filtering and searching output files, visualize the summaries of filtered data as well as exporting them. ChiRA is now part of the RNA workbench (RNA workbench).
A complete workflow for interactome data analysis is at https://usegalaxy.eu/u/videmp/w/rna-rna-interactome-analysis
If you want to look at already analyzed data, an example published history generated using the workflow on a CLEAR-CLIP dataset is available at https://rna.usegalaxy.eu/u/videmp/h/rna-rna-interactome-analysis
You can learn the usage of the ChiRA tool suite to analyze your interactome datasets from the Galaxy training material: RNA-RNA interactome data analysis.