Multi-omics Visualization Platform: An extensible Galaxy plug-in for multi-omics data visualization and exploration
a visualization plug-in that extends Galaxy-P’s advantages into the visualization of large, complex datasets
Proteogenomics, enables new insights into understanding the complex biological systems by integrating genomics/transcriptomics with mass spectrometry based proteomics data. It is a powerful approach to extract direct evidence of expression of novel protein sequences specific to a sample of interest. The importance of proteogenomics studies is reflected in the field of cancer and other genetic diseases.
The Galaxy for proteomics (Galaxy-P) team, has disseminated various tools along with a workflow necessary to perform proteogenomics analysis i.e. from raw data processing and sequence database generation to database searching and tools for interpreting the potential impact of identified sequence variants. Along with this, the Galaxy-P team has developed a complete proteogenomic informatics platform that combines genomics, transcriptomics and MS-based proteomics data. The Multi-omics Visualization Platform (MVP), a visualization plug-in, that extends Galaxy-P’s advantages into the visualization of large, complex datasets. This plug-in allows researchers to quickly inspect and verify the quality of their results as well offer an visulaization overview for deeper understanding of underlying spectral data.
MVP is primarily written in Javascript, with HTML5 and CSS to create the interactive user interface. It needs 3 separate SQLite databases as the primary input. The SQLite database schema helps in protein to peptide mapping. The MVP tool uses an SQlite schema to navigate between genome and the proteome.
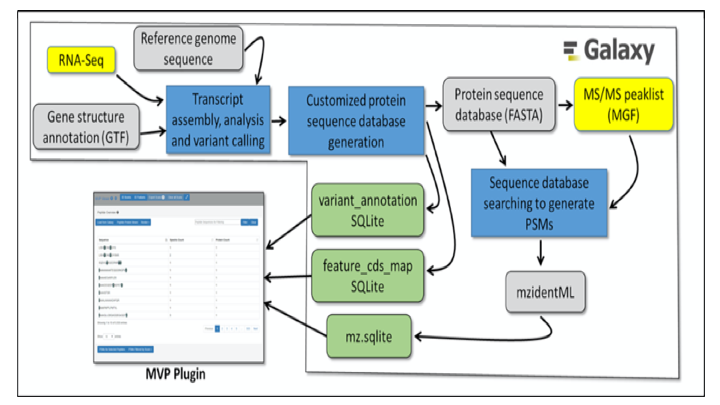
With the incorporation of Integrated Genomics Viewer(IGV) and Lorikeet, the MVP platform merges proteomic and genomic results into a single, accessible output. For large datasets, a user can easily navigate through the filter option and condense it down to a manageable subset making the database size not an issue. The MVP tool allows experimental results from either a peptide centric view, a PSM centric view (Lorikeet) or Protein centric view. The peptide centric view allows for rapid filtering based on sequences of interest. The PSM centric view allows for fast filtering of scores yielding the lowest decoy hit rate. And the protein centric view allows for an immediate examination of proteins of interest. This may be of particular interest to users who want to analyze variant proteins. The integrated IGV viewer offers interactive viewing of peptides mapped against the reference genome, plus an additional feature of incorporating tracks for standard-format sequence file (BAM, proBAM, BED).
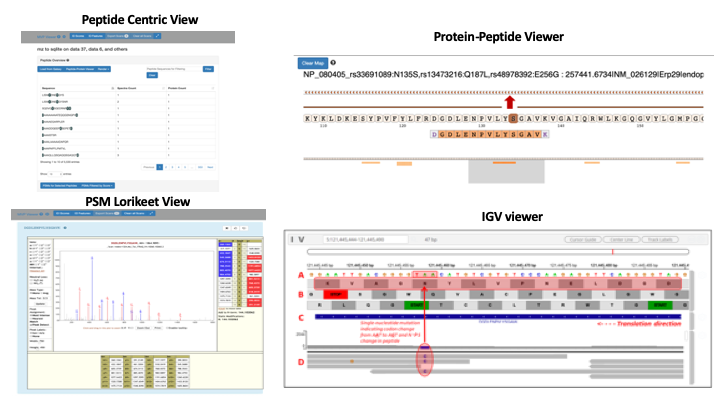
To learn more about how to use the MVP tool and its features, please refer the Galaxy training material.
Contributors
Galaxy-P @ University of Minnesota:
- Thomas McGowan
- James E. Johnson
- Ray Sajulga
- Praveen Kumar
- Subina Mehta
- Pratik Jagtap
- Timothy J. Griffin

