Omicron and SARS-CoV-2 genome surveillance
A first view of the Omicron lineage’s mutational pattern derived transparently and fully reproducibly from raw sequencing reads
The newly reported SARS-CoV-2 lineage B.1.1.529 made the headlines last week. On Friday then, the WHO declared the lineage a variant of concern (VOC) asking countries to, among other things, enhance surveillance and sequencing efforts to better understand circulating SARS-CoV-2 variants.
For several months, the Galaxy Project has kept running a free, public genome surveillance program based on raw sequencing data deposited in the public databases of the ENA and the SRA. Among the countries we have been collaborating with to ensure high-quality analysis of their national SARS-CoV-2 genome sequencing efforts has been South Africa, which we included recently in our public dashboard at https://covid19.galaxyproject.org/dashboard. With the first reports of B.1.1.529 appearing last week, we ran our analysis workflows on one of their latest batches of data, and can offer now a first view of the Omicron lineage's mutational pattern derived transparently and fully reproducibly from raw sequencing reads.
The full analysis history of a batch of 68 samples, most of which turn out to be of the Omicron lineage, enables everybody to verify claims made about the mutations found in the new VOC. No need to rely on hard to verify FASTA consensus sequences, nor on mainstream media - just check out the analysis and see the samples and the derived variant calls and lineage assignments right there.
For the impatient, here's the summary plot of the variant spectrum of the batch (taken directly from the linked history above):
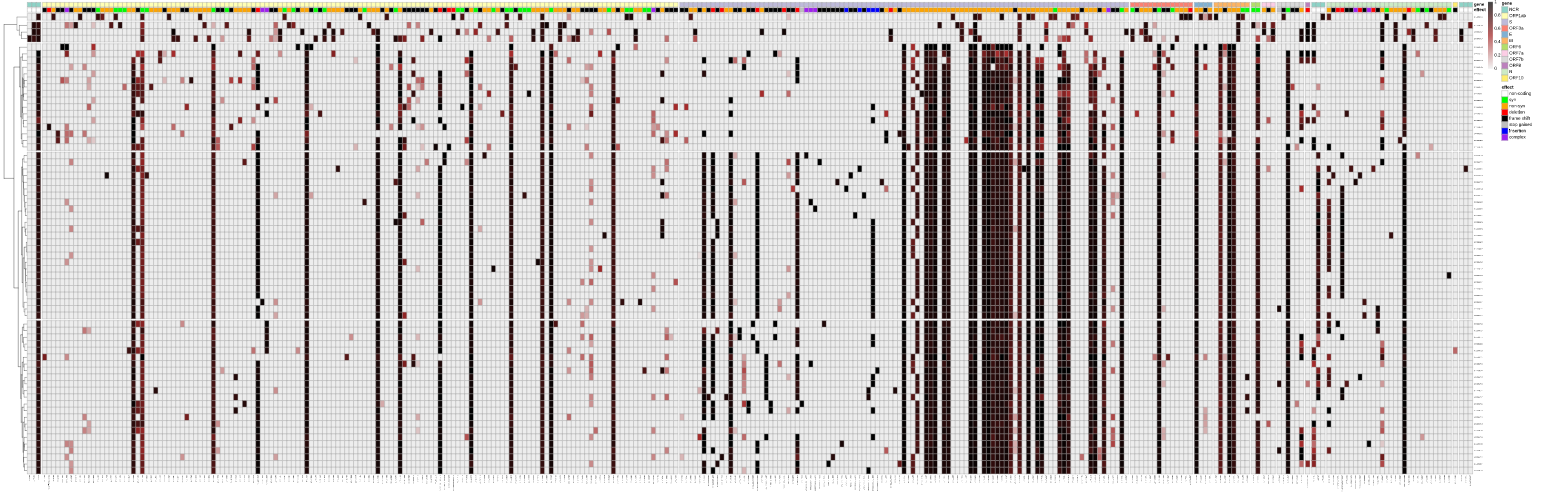
The reported, unprecedentedly high number of mutations in the spike protein (delineated by the narrow violet bar at the top of the plot) of the VOC can be identified clearly. The full analysis confirms all key mutations that have been publically reported.
A huge thank you goes to the KwaZulu-Natal Research Innovation and Sequencing Platform (KRISP) for submitting raw sequencing data of South African samples in an extremely timely manner under NCBI Bioproject PRJNA636748. It is open sharing of data of this kind that enables scientific insight (even if the information gathered is as unpleasant as a new VOC)!
In case you want to understand better how viral variant calling and lineage assignment work and even want to try things yourself, we have prepared a starting history with just the raw data and required accessory datasets for you to experiment with. Use it in conjunction with our dedicated training material to familiarize yourself with an aspect of science that has and, unfortunately, will continue to have a huge impact on our daily lives.
Update 2021-12-05
- We published the first selection analysis results about Omicron on virological.org: "Selection analysis identifies significant mutational changes in Omicron that are likely to influence both antibody neutralization and Spike function"
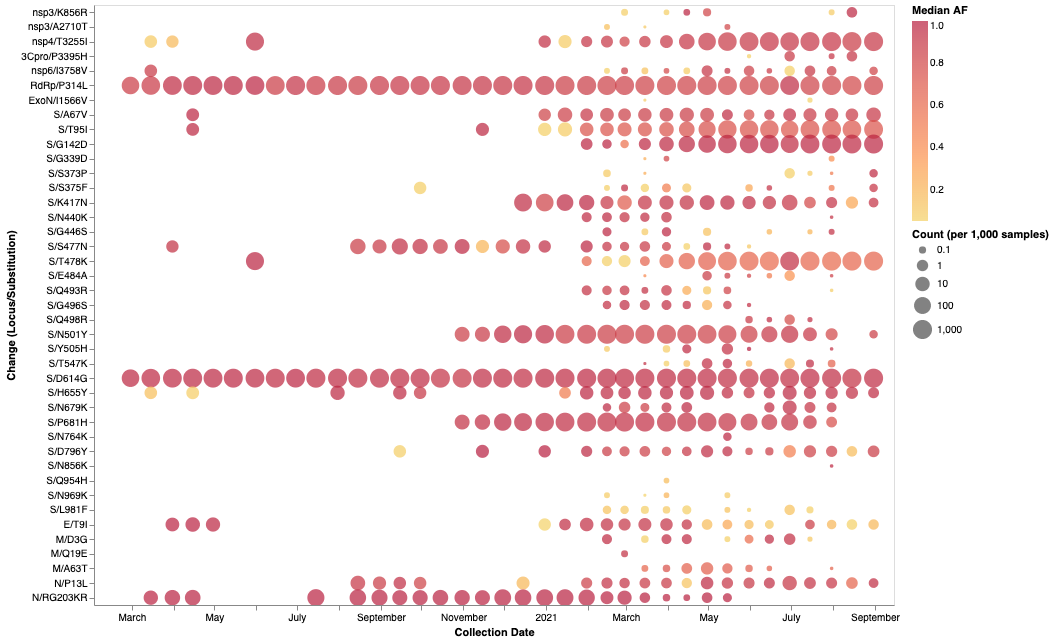
- You can see above the intra-host allelic variation seen at Omicron amino acid mutation sites in a subset of SARS-CoV-2 raw sequencing data since March 2020 analyzed using the Galaxy variant calling pipeline
- As you can see in the image, many Omicron mutations have been seen in subconsensus frequencies long before identification of Omicron.
- Conclusion: Intermediate frequency data is critical for early detection and you can get this data in a high quality from us. Happy research!
Update 2021-12-08
New analysis results are available from both ONT and Illumina data:
- Galaxy History - omicron-ont-midnight-batch-1 (141 ONT runs, 130 assigned to B.1.1.529)
- Galaxy History - omicron-illumina-articv4-batch-1 (70 Illumina runs, 66 assigned to B.1.1.529)
Further readings:




