Galaxy Community Conference - a brief summary!
A brief summary
The 2019 Galaxy Community Conference (GCC2019) was held in Freiburg, Germany, 1-8 July. 2019 was the largest GCC ever with 231 participants, 39 Galaxy training courses, 65 talks, 54 posters, 13 demos, and more than 30 different BoFs. Our participants came from 27 different countries of Eastern- and Western Europe, North America, Asia, Oceania and Sub-saharan Africa.


GCC2019 offered several new features in the schedule. We still started with a day of dual-track training and ended with up to four days of CollaborationFest, but the main meeting:
- Expanded from two days to three days.
- Offered dual presentation tracks most of the time
- Intermixed training and presentation sessions.
These changes supported the record amount of content that was offered this year and they made the meeting more interactive than in past years. The meeting featured plenary & parallel sessions; invited, accepted, and lightning talks; poster and demo sessions; birds-of-a-feather meetups; social events; and training sessions every day.

Have a look at our conference book to get all information.
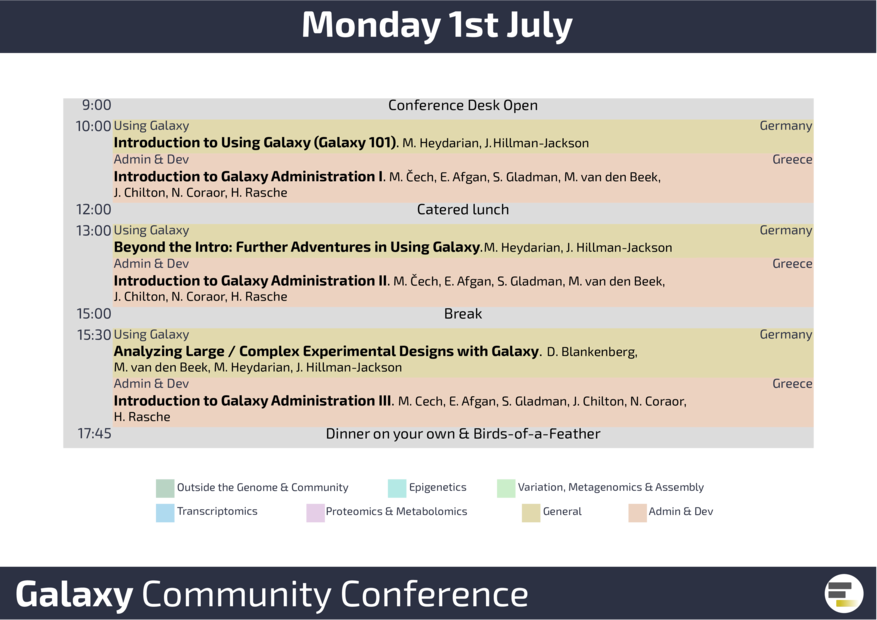
GCC2019 started on Monday, July 1st, with a dedicated training day for Galaxy beginners offering a user-centric track with “Introduction to using Galaxy” in parallel to the admin and developer focused “Introduction to Galaxy Administration”. Participants interested in long read sequencing and wet-lab work could visit Nanopore loading zone and try the Oxford Nanopore MinION. In the evening, the first Birds-of-a-Feather (BoFs) were offered for attendees interested in Single Cell data analysis, Genome Annotation, and Computational Chemistry.
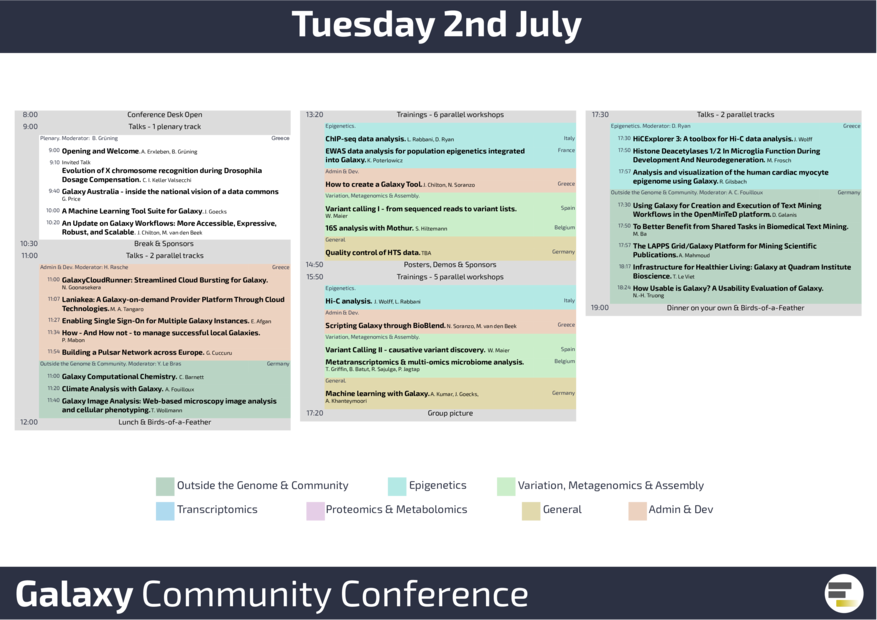
GCC2019 was officially opened on Tuesday, July 2nd, by Björn Grüning and Anika Erxleben. One of our Galaxy power users, Claudia Isabelle Keller Valsecchi, was invited to present her impressive data on “Evolution of X chromosome recognition during Drosophila Dosage Compensation". After several talks on Galaxy Australia, Machine learning, and Galaxy Workflows, talks and workshops were offered in two tracks, one more user-driven, the other more admin-related. Galaxy is used across so many fields of research, as we heard e.g. from Anne Fouilloux talking about Climate analysis with Galaxy. During the afternoon break, the participants could discover interesting Galaxy projects in the poster and demo session. The Epigenetics sessions showed how to perform 16S analysis with mothur, ChIPseq data analysis and HiC data analysis. Beside multi-omics data analysis in Galaxy, Machine learning with Galaxy was presented for the first time.
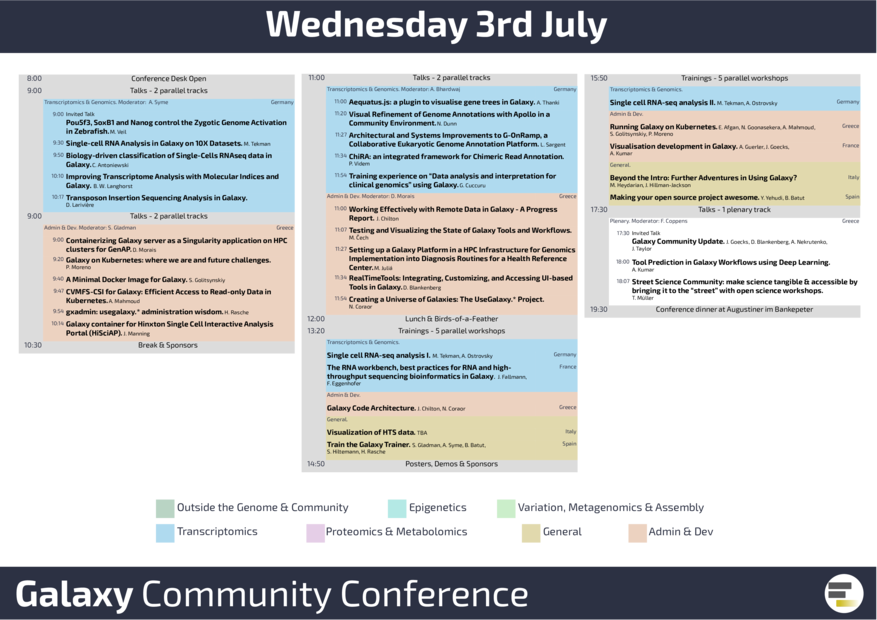
Wednesday's Galaxy user track was dedicated to transcriptomics and genomics data analysis. The demand for single cell RNA-seq data analysis was very obvious in the presentations by Mehmet, Christophe, and Jonathan. Running Galaxy for annotation with e.g. Apollo, G-ONRamp, and ChiRA was successfully shown in talks, posters, trainings, and demos. Galaxy also now runs on Kubernetes. Before the conference dinner, a public science project was presented by the Street Science Community. More than 150 people joined us for the conference dinner in Augustiner im Bankepeter. We spent an amazing evening in the beer garden with our friends and colleagues. Thank you all!
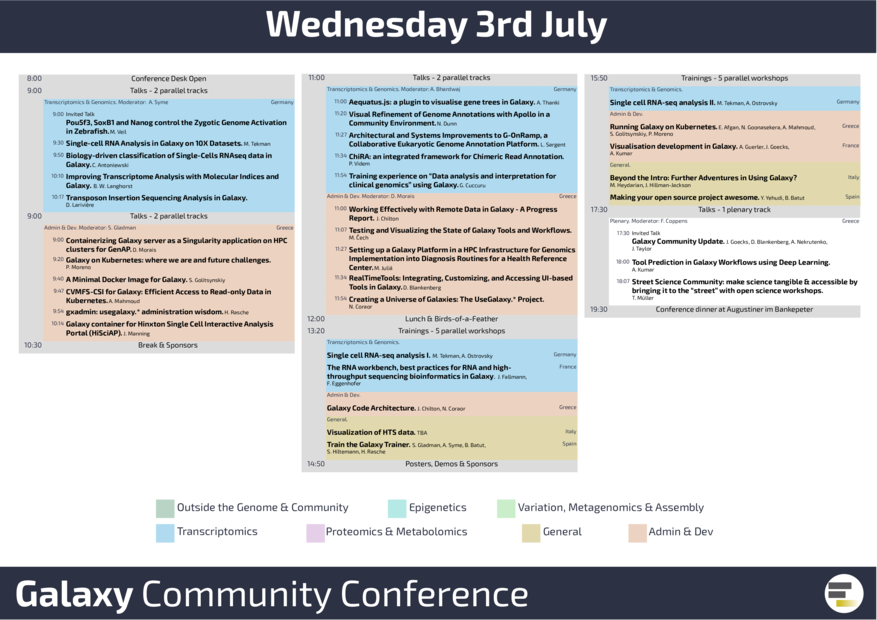
Proteomics, metabolomics, metagenomics, and variant calling data analysis were presented on Thursday in various talks, workshops, demos, and posters, such as microbiome data, ProteoRE, IRIDA, and Workflow4Metabolomics. With Galaxy-E Yvan brought our “Wonderful World” a bit closer, he presented the Galaxy Ecology project with new, dedicated Galaxy Europe subdomain. Before closing the conference, Bérénice gave an update on the Galaxy training network.
The GCC2019 core conference days were followed by 4 days of CollaborationFest. During these 4 days ~100 people worked on library enhancements, Galaxy packages, development of an ATAC-seq tutorial, support for select parameters in workflows, tool menu style, EDAM in Galaxy, Training development kit, and on a lot of work on usegalaxy.*.
Freiburg Challenge
A great opportunity to get to know the beautiful city of Freiburg was taking part in the Freiburg challenge. Hereby we ask the participants to find or do something in Freiburg and prove it by taking a picture. Many participants had lots of fun and successfully had e.g. a swing on the Dreisam, walked into a Bächle to cool down, did the "I am the king of the world" pose on top of the Schlossbergturm ot ate a Lange Rote at the Münsterplatz. The winners were awarded by a typical Freiburg Bächleboat.

Thank you: Scientific Program Committee
- Matthias Bernt, Helmholtz Centre for Environmental Research – UFZ
- Dan Blankenberg, Cleveland Clinic Lerner Research Institute
- Gildas Le Corguillé, Station Biologique de Roscoff
- Maria Doyle, Peter MacCallum Cancer Centre
- Anika Erxleben, University of Freiburg
- Melanie Föll, University of Freiburg
- Anne Claire Fouilloux, University of Oslo
- Carrie Ganote, Indiana University
- Jeremy Goecks, Oregon Health & Science University
- Björn Grüning, University of Freiburg
- Jennifer Hillman-Jackson, Penn State University
- Saskia Hiltemann, ErasmusMC
- Hans-Rudolf Hotz, Friedrich Miescher Institute for Biomedical Research
- Nancy Ide, Vassar College
- Pratik Jagtap, University of Minnesota
- Yvan Le Bras, Muséum national d’histoire naturelle
- Thomas Manke, Max Planck Institute of Immunobiology and Epigenetics
- Pablo Moreno, EMBL-EBI
- Anton Nekrutenko, Penn State University
- Krzysztof Poterlowicz, University of Bradford
- Helena Rasche, University of Freiburg
- Devon Ryan, Max Planck Institute of Immunobiology and Epigenetics
- Beata Scholz, University of Debrecen
- Nicola Soranzo, Earlham Institute
- Peter van Heusden, University of the Western Cape, South African National Bioinformatics Institute (SANBI)
- Ralf Weber, University of Birmingham
Thank you: Organizers
Conference
- Bérénice Batut, University of Freiburg
- Dave Clements, Johns Hopkins University
- Frederik Coppens, VIB, UGent, ELIXIR Belgium
- Monika Degen-Hellmuth, University of Freiburg
- Rolf Backofen, University of Freiburg
- Anika Erxleben, University of Freiburg
- Björn Grüning, University of Freiburg
- Mo Heydarian, Johns Hopkins University
- Helena Rasche, University of Freiburg
- Devon Ryan, Max Planck Institute of Immunobiology and Epigenetics
- Jenn Vessio, Johns Hopkins University
CoFest
- Dannon Baker, Johns Hopkins University
- Frederik Coppens, VIB and ELIXIR Belgium
- Maria Doyle, Peter MacCallum Cancer Centre
- Mo Heydarian, Johns Hopkins University
- Jennifer Hillman-Jackson, Penn State University
- Brad Langhorst, New England Biolabs
- Marius van den Beek, Institut Curie