New Paper "Survey of metaproteomics software tools for functional microbiome analysis"
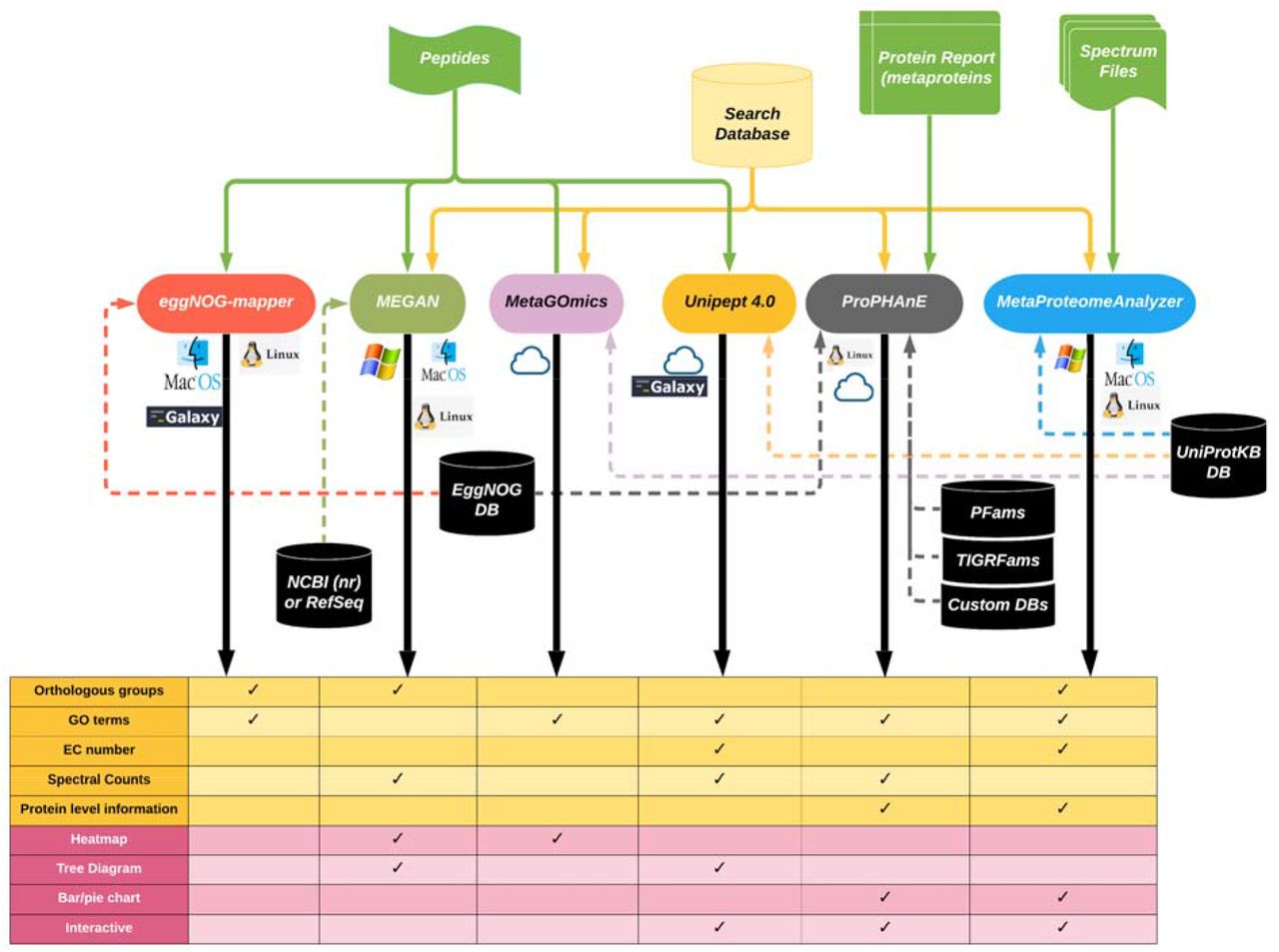
Our metaproteomics and microbiomics communities are constantly growing. This article on bioRxiv may be interesting for many of you: "Survey of metaproteomics software tools for functional microbiome analysis". The authors compare six open software tools for microbiome analysis, including Unipept 4.0 and eggNOG-mapper, which are already installed in the European Galaxy server.
Abstract - To gain a fuller appreciation of microbiome dynamics, researchers characterize the functional role of expressed microbial genes/proteins. This can be accomplished through metaproteomics, which characterizes the protein complement of the microbiome. Several software tools exist for analyzing microbiomes at the functional level by measuring their combined proteome-level response to environmental perturbations. In this survey, we evaluate the performance of several available tools, so that researchers can make informed decisions regarding software choice based on their research goals.
As representative data for this evaluation, mass spectrometry-based proteomic data was generated from plaque samples for dental caries and grown with and without sucrose in paired biofilm reactors. Microbial peptides identified after search of tandem mass spectrometry data from one sample pair were subjected to functional analysis using software tools such as eggNOG-mapper, MEGAN6, MetaGOmics, MetaProteomeAnalyzer (MPA), ProPHAnE, and Unipept to generate Gene Ontology (GO) terms for functional annotation.
Among these software tools, notable differences in functional annotation were detected after comparing differentially expressed protein functional groups. Based on the generated GO terms of these tools we performed a peptide-level comparison to evaluate the quality of their functional annotations. A BLAST analysis against Universal Protein Knowledgebase revealed that the sensitivity and specificity of functional annotation differed between tools. For example, eggnog-mapper mapped to most number of GO terms, while Unipept generated the most precise GO terms. Based on our evaluation, metaproteomics researchers can choose the software according to their analytical needs and developers can use the resulting feedback to further optimize their algorithms. To make more of these tools accessible via scalable metaproteomics workflows, eggNOG-mapper and Unipept 4.0 were incorporated into the Galaxy platform.