Galaxy at JOBIM
The JOBIM conference, the annual event of the French bioinformatics and biostatistic community, brought together researchers, educators, and innovators in bioinformatics to discuss cutting-edge tools, workflows, and collaborative efforts. Among the highlights of this year's event was the significant presence of Galaxy. This blog post explores the key contributions, presentations, and innovations centered around Galaxy at JOBIM 2025.
Keynote: From Genomes to Galaxies—The Evolution of Scalable Open Data Analysis
The keynote address by Björn, on behalf of the Global Galaxy community, was highly appreciated. Björn provided a compelling overview of how Galaxy has evolved over two decades into a scalable and accessible platform for open data analysis. From genomics to astronomy, Galaxy’s versatility has enabled researchers worldwide to tackle complex computational challenges while fostering collaboration and reproducibility.

Björn emphasized that Galaxy's success lies in its ability to empower both novice users and advanced bioinformaticians. Its user-friendly interface, extensive library of tools, and commitment to open science have made it a go-to platform for diverse scientific communities, including 'omics, ecology and astronomy.
Contributions from the French Galaxy Working Group
The French Galaxy Working Group animated by the IFB/ELIXIR-FR showcased the breadth of their contributions through several posters and presentations:
-
"Exploring the Richness of the French Galaxy Ecosystem" [Poster]
Bérénice, Gildas, and Anthony presented a detailed analysis of the French Galaxy ecosystem. Their work underscored the diversity of tools, workflows, and community-driven initiatives that have made France a hub for Galaxy innovation.
-
"Breaking Myths: The Reality of Galaxy's Capabilities and Impact" [Poster]
Gildas, Anthony, Björn, and Bérénice addressed common misconceptions about Galaxy. The poster highlighted how the platform is not just a tool for data analysis but also a powerful enabler of reproducible research, community-driven development, and educational outreach.
-
"Galaxy Training: A powerful framework for teaching!" [Poster]
Bérénice and Björn demonstrated how Galaxy serves as an invaluable resource for teaching bioinformatics, especially via the Galaxy Training Network. The platform's interactive environment allows educators to create engaging workflows that teach students about data analysis, reproducibility, and best practices in computational biology.
-
"Genome Annotation Tooling in Galaxy" [Poster]
Romane and Anthony highlighted contributions from the EuroScienceGateway project to improve genome annotation workflows within Galaxy. These advancements enhance the platform's utility for researchers studying genomics and evolutionary biology.
-
"Galaxy as a Service (GaaS)" [Poster]
Rémi and Juliette introduced GaaS, a TypeScript toolkit designed for developers who want to build custom web applications powered by Galaxy. This innovation bridges the gap between Galaxy's robust analysis capabilities and modern web development practices.
UseGalaxy.fr was also highlighted by Anthony and Bérénice in the IFB presentation during the Network Presentations session.




ABRomics: Antibiotic Resistance Surveillance and Resarch with Galaxy
ABRomics, a project centered on antibiotic resistance (AMR) surveillance, was another highlight of the conference:
-
"ABRomics-analysis: a web service for the analysis of Antibiotic Resistance in bacterial genomes" [Demo]
Amanda demonstrated ABRomics-analysis, a web service that leverages Galaxy to analyze bacterial genomes and monitor the spread of antibiotic resistance genes. This tool is critical for identifying and mitigating public health threats posed by multidrug-resistant bacteria.
-
Hugo and colleagues showcased how ABRomics is using Galaxy workflows to develop metagenomic pipelines for national AMR surveillance. Their work emphasizes the importance of scalable, reproducible bioinformatics solutions in addressing global health challenges.
Mention of Galaxy in Other Projects
Galaxy's versatility was evident in its mention across various projects:
-
"Fast answers to simple bioinformatics needs and capacity building in an island context, a focus on microbial omics data analysis" [Talk]
This talk by David Couvin emphasized the use of Galaxy for training purposes, particularly in an island context, focusing on microbial omics data analysis
-
"Met4J: a library, a toolbox and a workflow suite for graph-based analysis of metabolic networks" [Talk]
The Met4J tools were showcased as a workflow suite and available as tools in Galaxy, facilitating graph-based analysis of metabolic networks.
-
"Madbot, a metadata and data brokering online tool to ensure the adoption of standards and FAIR principals in an open science context" [Talk]
Madbot was presented as an online tool that connects with Galaxy, alongside platforms like ENA and OMERO, to manage and share scientific data, promoting open science standards.
-
"Ten years of the Pasteur's Bioinformatics and Biostatistics Hub: achievements and perspectives" [Talk]
The Pasteur hub highlighted its work including its Galaxy instance, designed to support community bioinformatics analyses and contributions to ELIXIR's tool platform.
-
"vvv2_display: Galaxy Workflows for Variant Calling and Annotation" [Poster]
Alexandre Flageul and colleagues highlighted how Galaxy workflows are being used to compute, summarize, and visualize variant calling and annotations in viral genomes. Tools like vvv2_align_SE and vvv2_display are available on the Galaxy Toolshed, making them accessible to a wide audience.
-
"SIDURI: A User-Friendly Portal for Fermentation Data Analysis" [Poster]
The SIDURI project integrates Galaxy into its workflow management system, enabling users to analyze fermentation data through an intuitive interface. This integration demonstrates Galaxy's adaptability as a platform for diverse scientific domains.
-
"IFB/ELIXIR-FR contributions to the ELIXIR Training Platform" [Poster]
French contributions to the ELIXIR Training Platform were also highlighted, with a focus on the Galaxy Training Network (GTN). These efforts underscore the importance of training and education in fostering a skilled bioinformatics workforce.
-
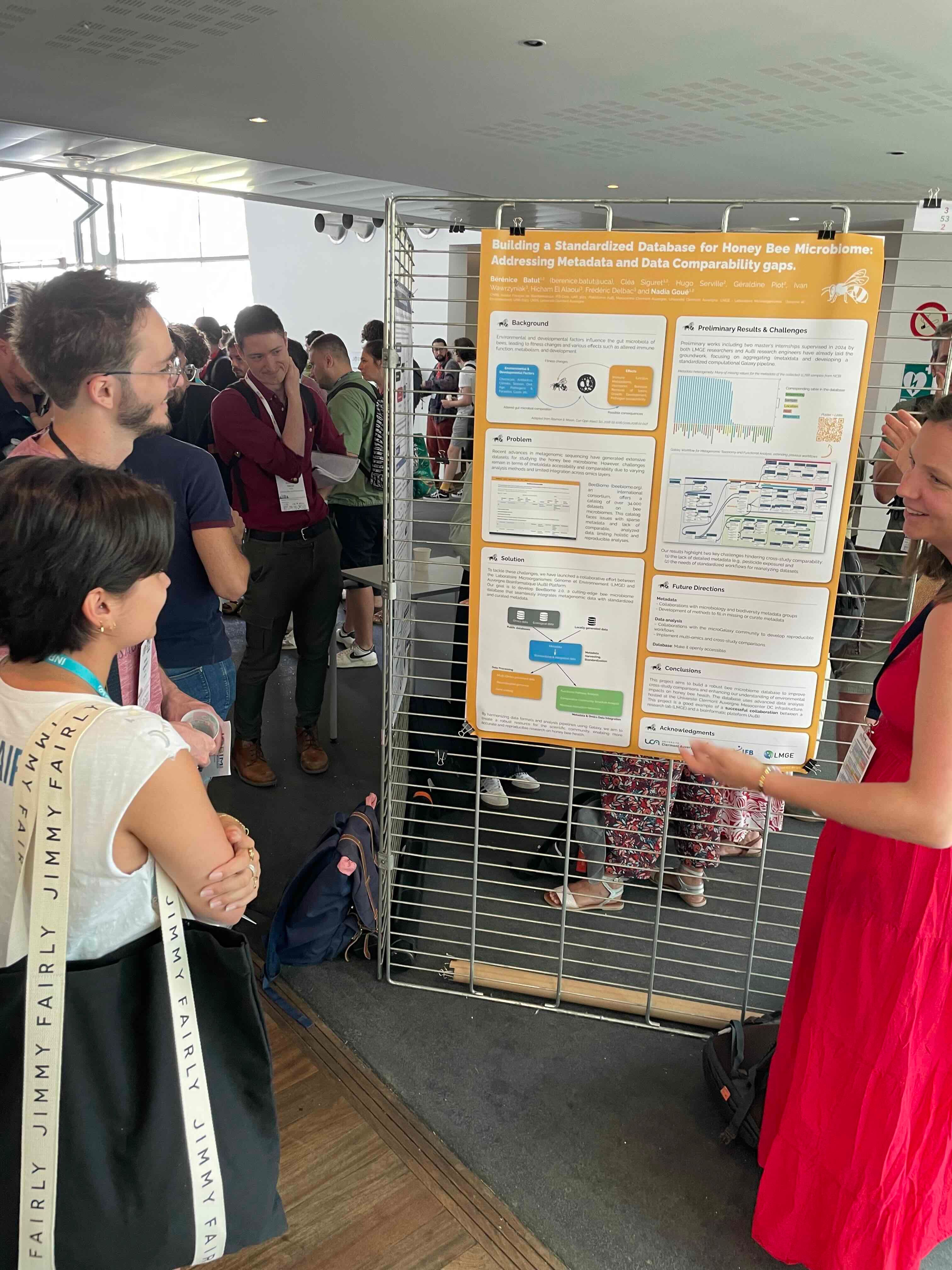
Bérénice and Nadia presented collaborative work between AuBi platform and the Host-Parasite team at LMGE, focusing on data comparability and metadata quality in microbiome research. Preliminary efforts included aggregating metadata and developing standardized Galaxy pipelines. Key challenges identified were the lack of detailed metadata and standardized workflows for dataset reanalysis. The project aims to build a robust bee microbiome database using advanced data analysis.
Conclusion
JOBIM 2025 was a testament to the enduring impact and versatility of Galaxy as a platform for open data analysis. From keynote addresses to posters, demo, and talk, the conference showcased how Galaxy continues to empower researchers, educators, and innovators across diverse fields. As the global community of Galaxy users grows, its role in advancing reproducible science, supporting education, and addressing real-world challenges like antibiotic resistance will only continue to expand.
Galaxy's journey from genomics to galaxies is far from over—it remains a beacon of innovation and collaboration in bioinformatics.

