An update on Galaxy Genome Annotation
Galaxy as a platform for the annotation of genomes
Galaxy Genome Annotation
As presented at GCC2023 and EGD2023, a lot of new exciting developments have been made in Galaxy for the annotation of genomes!
This has been done in the frame of the GGA community of practice that you are really welcome to join if you have interest in genome annotation within Galaxy (as a user, developer, trainer, …).
New tools!
We’ve worked hard to integrate into Galaxy many state-of-the-art genome annotation tools! Here’s the complete, categorized, list:
Repeated elements detection
| Tool | Version | UseGalaxy.eu | |---|---|---|---|---| | RepeatModeler | 2.0.4 | link | | RepeatMasker | 4.1.5 | link | | Red | 2018.09.10 | link |
Gene detection for prokaryotes
| Tool | Version | UseGalaxy.eu | |---|---|---|---|---| | Prokka | 1.14.6 | link | | Bakta | 1.8.2 | link |
Gene detection for eukaryotes
| Tool | Version | UseGalaxy.eu | |---|---|---|---|---| | BRAKER3 | 3.0.3 | link | | BRAKER2 | 2.1.6 | link | | Helixer | 0.3.2 | link | | Funannotate | 1.8.15 | link | | MAKER | 2.31.11 | link | | TSEBRA | 1.1.1 | Available soon |
Functional annotation
| Tool | Version | UseGalaxy.eu | |---|---|---|---|---| | Antismash | 6.1.1 | link | | InterProScan | 5.59-91 | link | | EggNOG-Mapper | 2.1.8 | link | | DeepSig | 1.2.5 | link |
Evaluation methods
| Tool | Version | UseGalaxy.eu | |---|---|---|---|---| | Genome annotation statistics | 0.8.4 | link | | BUSCO | 5.4.6 | link |
Annotation comparison
| Tool | Version | UseGalaxy.eu | |---|---|---|---|---| | AEGeAN | 0.16.0 | link | | Funannotate compare | 1.8.15 | link |
General alignment tools
| Tool | Version | UseGalaxy.eu | |---|---|---|---|---| | BLAST | 2.14.1 | link | | Exonerate | 2.4.0 | link | | Diamond | 2.0.15 | link | | Miniprot | 0.12 | link |
Long non-coding RNA prediction
| Tool | Version | UseGalaxy.eu | |---|---|---|---|---| | FEELnc | 0.2.1 | link |
Visualisation
| Tool | Version | UseGalaxy.eu | |---|---|---|---|---| | JBrowse | 1.16.11 | link | | Circos | 0.69.8 | link | | GeneNoteBook | 0.4.8 | link |
New workflows!
Over the last years we have collected a lot of different workflows for different use-cases. All the GTN tutorials below, for example, are also available as workflows. However, they might be hard to find and as a consequence people can be tempted to redo them. To fix that we have started to push our workflows to the Intergalactic Workflow Commission, adding workflow tests and pushing them to workflowhub.eu and Dockstore for broader dissemination.
You can see the first IWC workflow from the GGA community at workflowhub or on GitHub as part of the IWC organisation.
Next up is the functional annotation workflow and our GTN workflows - see below.
New visualisations!
Galaxy is much more than just a workflow system and one area that we are trying to improve is Visualisation!
Check out our tutorials about this:
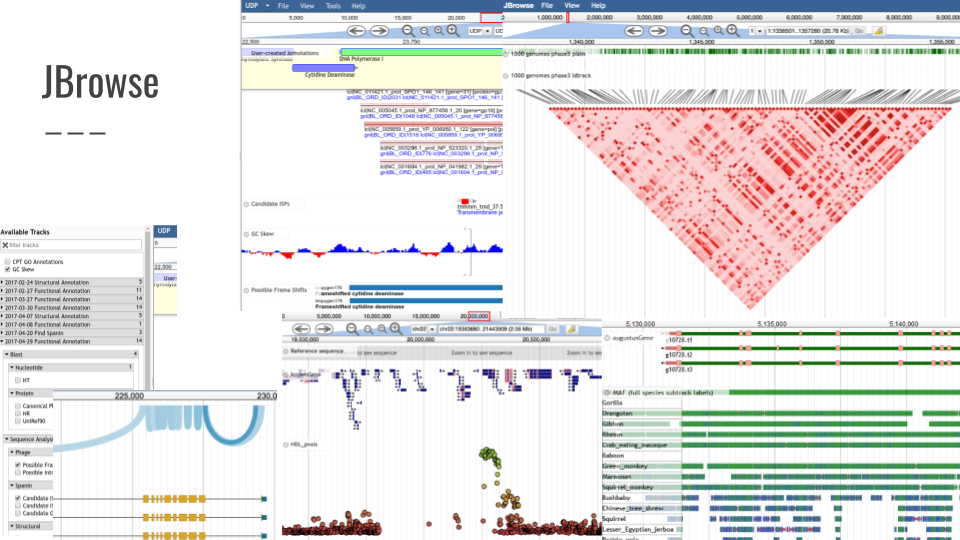
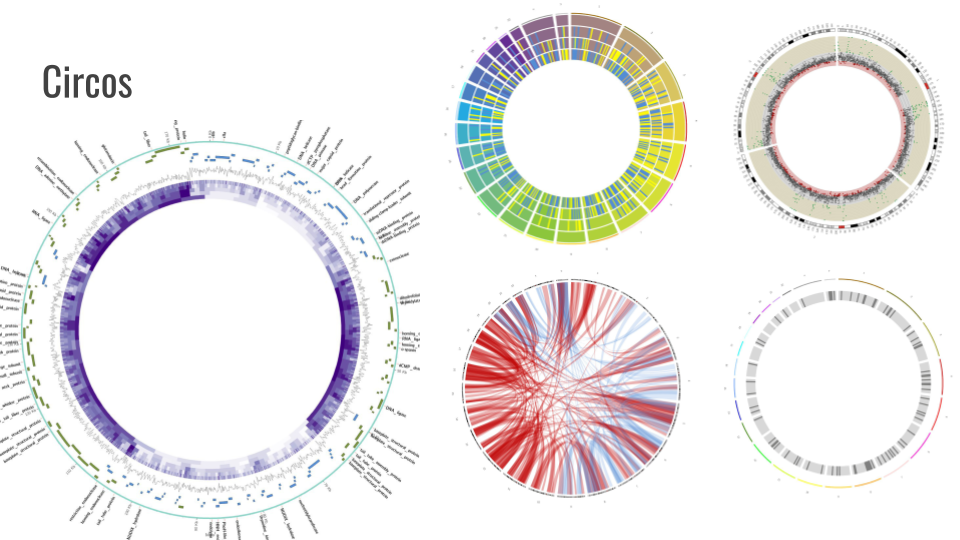
We also added support for GeneNoteBook: a nice way to present functional annotation data into an easy-to-navigate website:
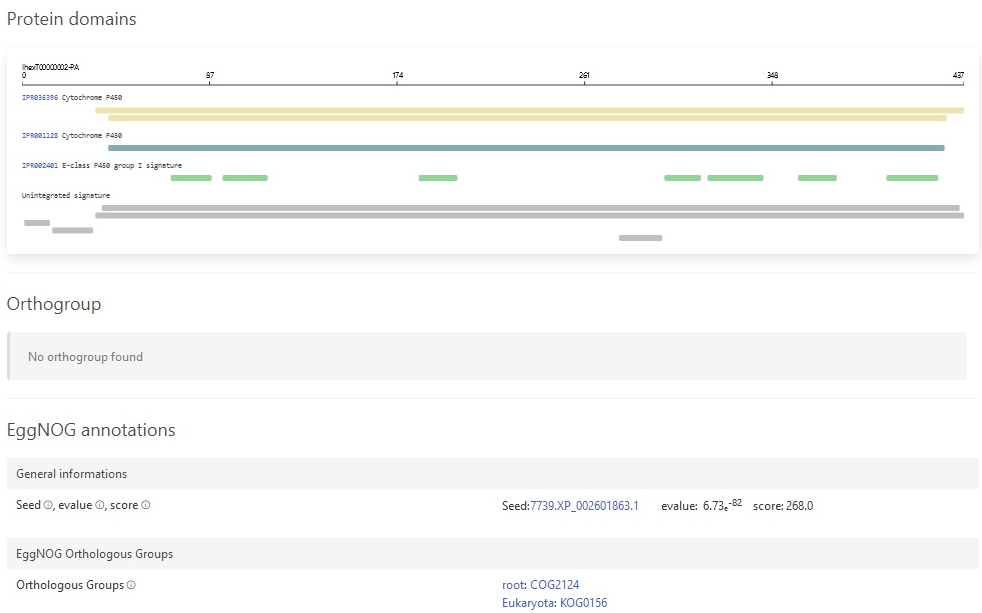
New and updated training material!
We have updated and added new training material for everyone that wants to learn Genome Annotation. If you don’t know where to start check out our recommended learning path and learn from “Introduction to Genome Annotation” to “Refining Genome Annotations with Apollo” everything that you need to know about Genome Annotation:
Alternatively, you can jump right into you favorite topic:
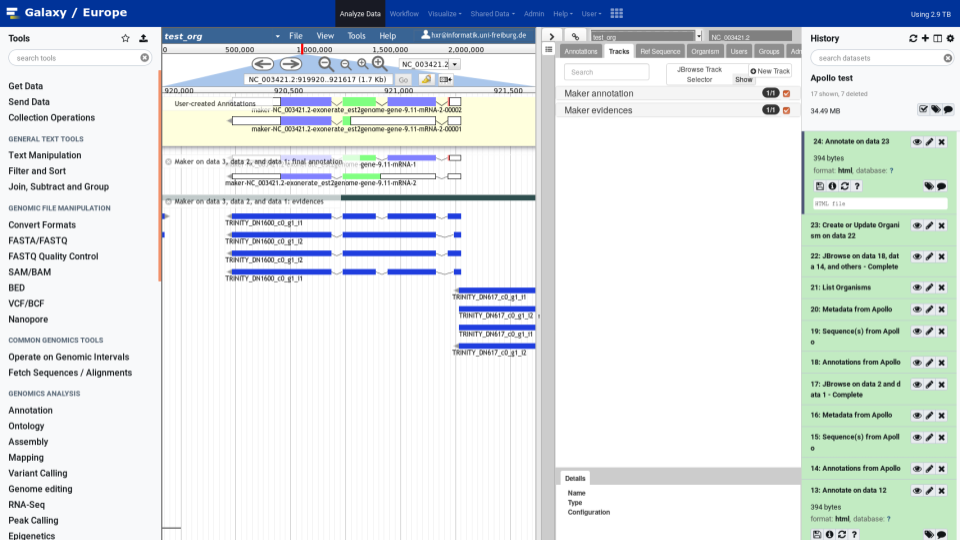
Manual curation and collaborative annotation made easy
Apollo is a web service for Collaborative Annotation - a “Google Docs of Genome Annotation” if you like to think in Google Terms. It allows for real-time, collaborative genome annotation, editing, and review of your favorite genome. Genome Annotation requires manual curation and review many hands are needed - Biology is a team sport! And Apollo is a wonderful tool to support your team.
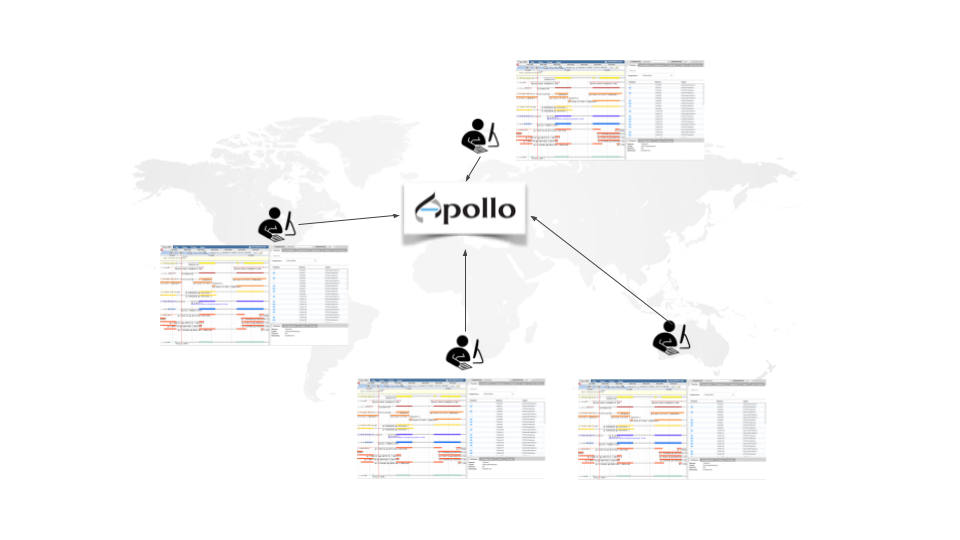
To make your life easy we worked closely with the Apollo team and bridged Galaxy with Apollo. You can do automatic annotation with Galaxy, using its powerful workflow system, and afterwards send the data to Apollo for manual curation and collaborative editing! A typical Galaxy-Apollo use-case might look like that:
- Fetch Data
- Analyse in Galaxy
- Send to Apollo
- Collaboratively Annotate → repeat
The Earth BioGenome Project
Building on the Assembly workflows that have been developed as part of the VGP/BGE/ERGA (→ in short EBP) we are planning to deliver high-quality workflows and interactive tools for your genome annotation efforts. Ideally, running assembly and annnotation workflows after each other 😊
What’s next?
Any tool or workflow missing in our catalog? Please tell us and we will do our best to make it available!
We already have these items on our TODO list, stay tuned:
- JBrowse2
- GALBA
- OMark
- BUSCO update
- InterProScan update
Join us on our chat channel to keep in touch!
