Shiny phyloseq interactive tool available on usegalaxy.eu - practical outcome of the Bioconductor Hackathon
We are thrilled to announce, that thanks to the help of the Bioconductor community, we could deploy the Shiny phyloseq App as an interactive tool (IT) on usegalaxy.eu
Shiny phyloseq interactive tool (IT)
Shiny apps are web apps using R functionality, that give easy responsive access to R packages. Interactive tools are a great way to work with data interactively and responsive using Galaxy. In theory, all shiny apps could become ITs, but so far the wrapping and deployment of shiny apps as ITs was technically challenging. To facilitate the integration of Shiny apps as ITs, the Freiburg Galaxy team, with the support of the de.KCD project, conducted a two-day hackathon in February, collaborating with members from the Bioconductor community, including Charlotte Soneson, Hans-Rudolf Hotz, and Federico Marini. During the event, best practices for developing ITs using Shiny apps were established, with a primary focus on creating a Docker image that can serve as a starting point for adding any Shiny app. As a proof-of-concept the shiny-phyloseq app has been wrapped as IT. A fork of this Docker image tailored for the shiny app is available here: https://github.com/paulzierep/docker-phyloseq. The image can be run locally to test the app and then must be deployed to quay.io or any other Docker registry. This app allows to perform dynamic analysis of metabarcoding/amplicon data such as:
- filter data based on metadata and taxonomy
- plot alpha diversity
- plot distance networks
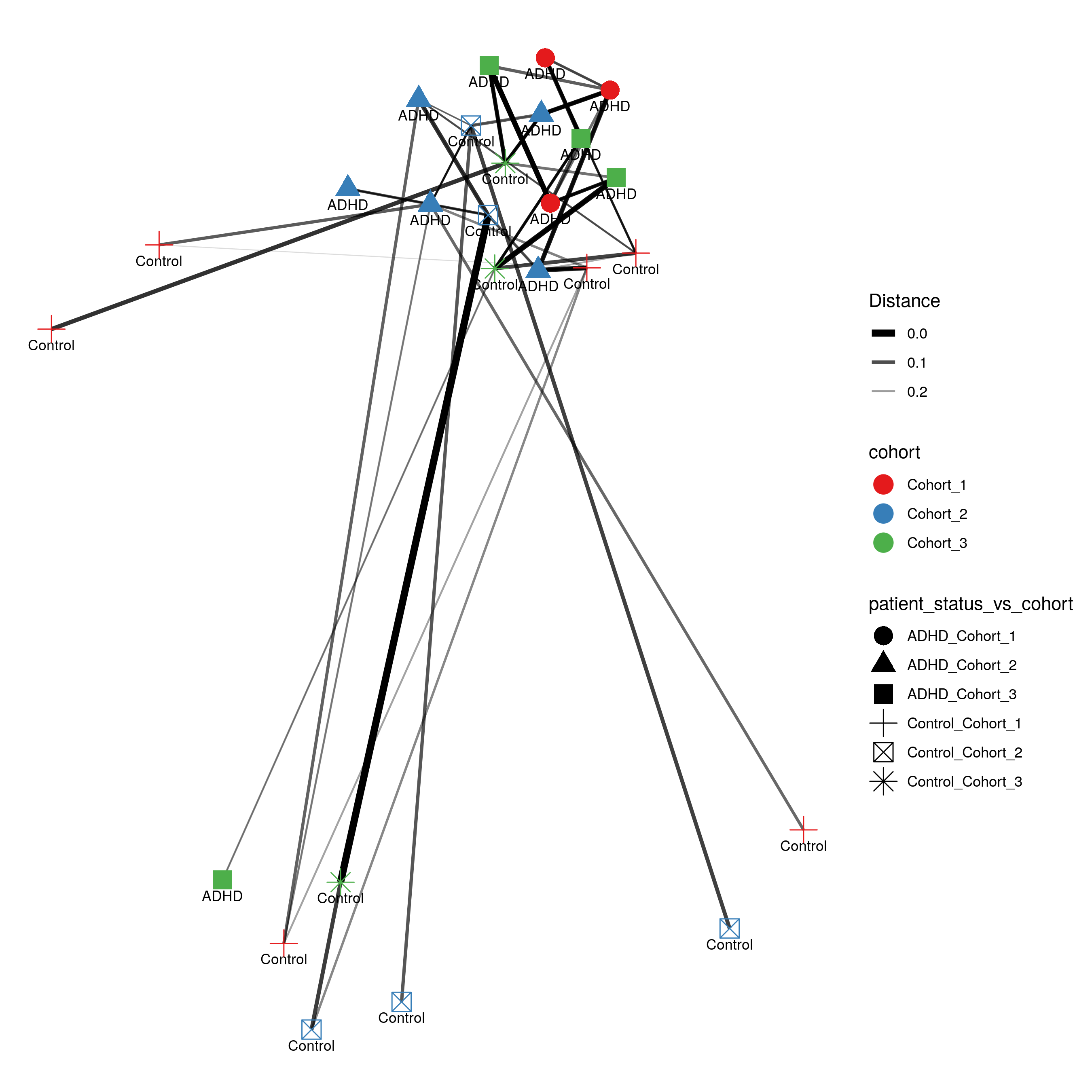
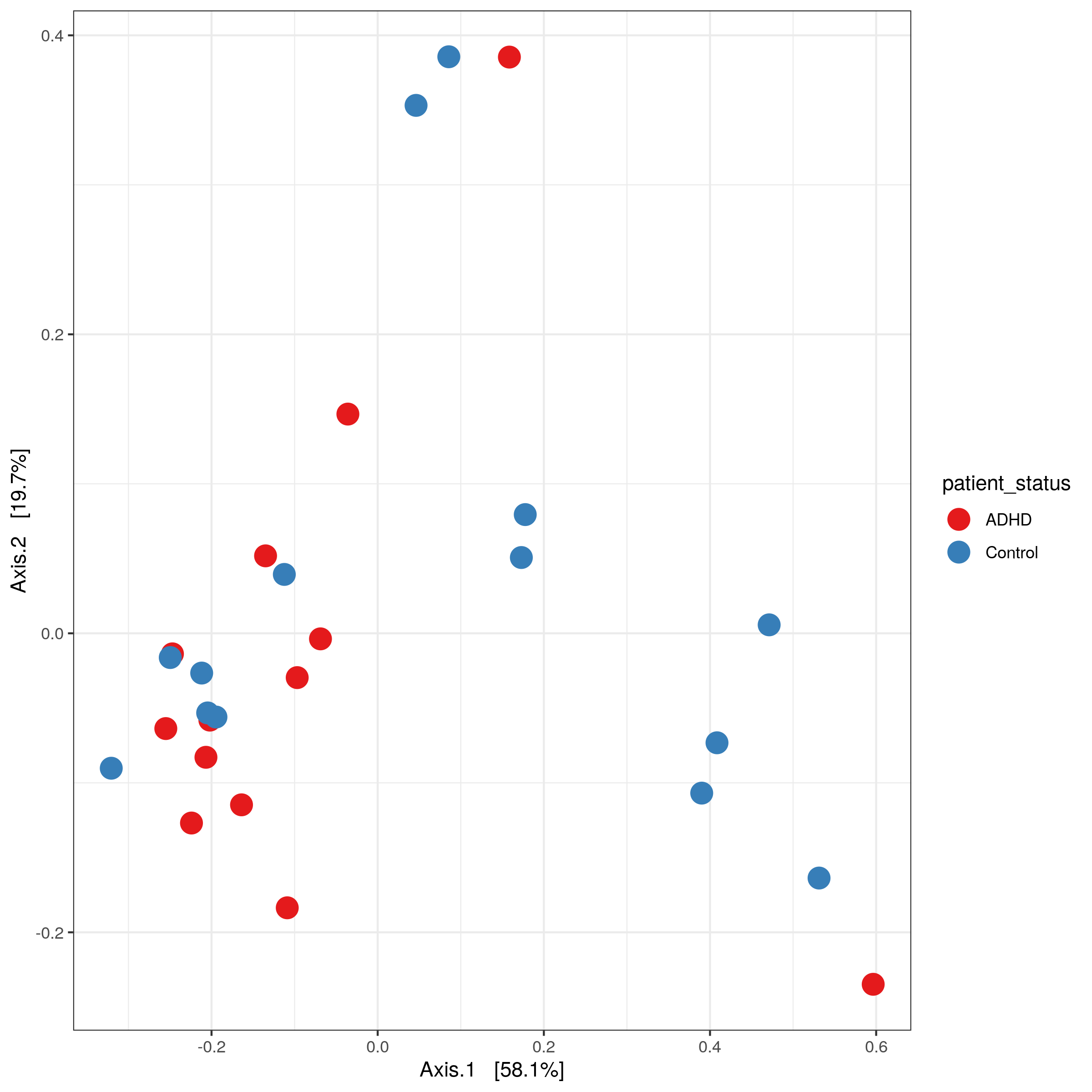
- ordination plots
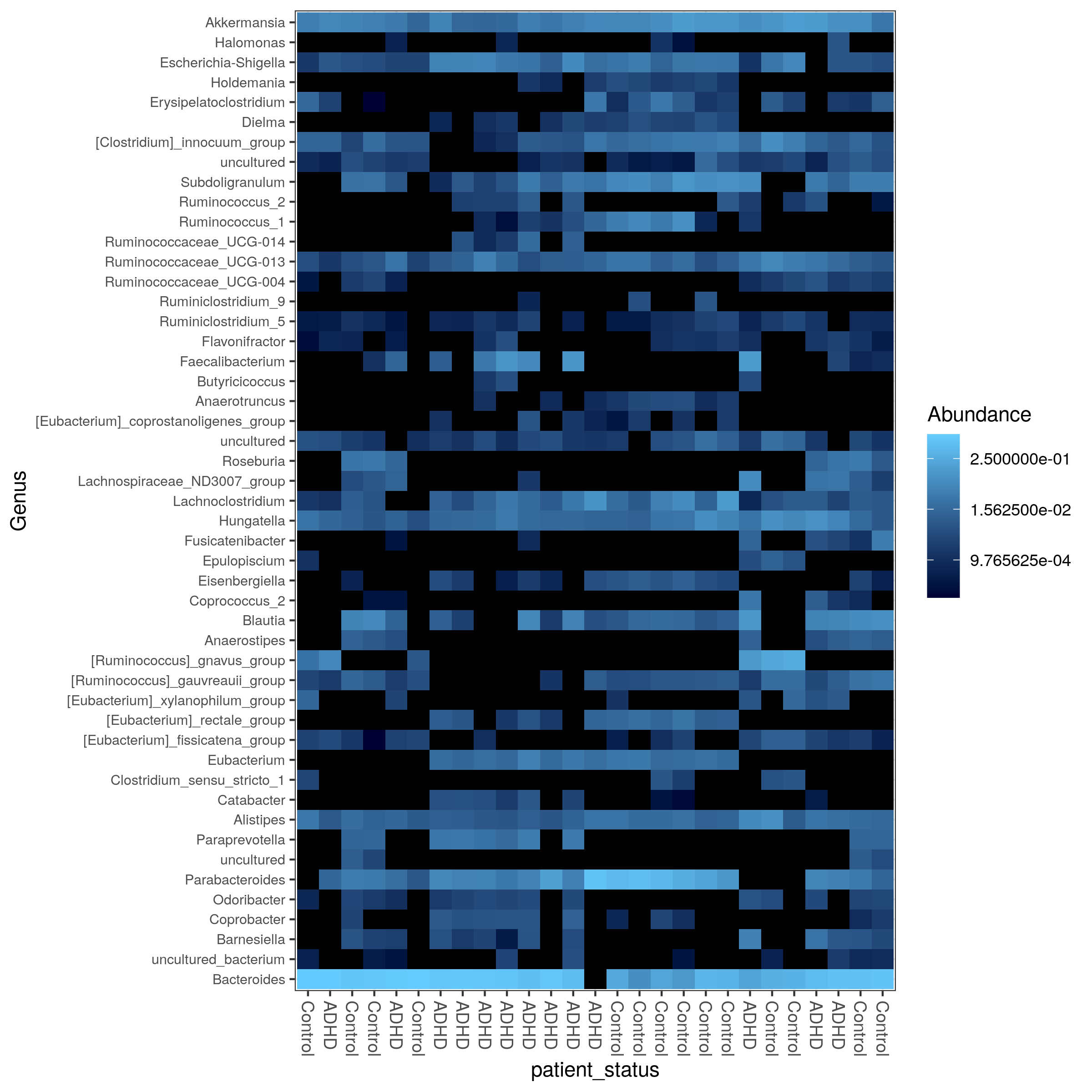
- heatmaps
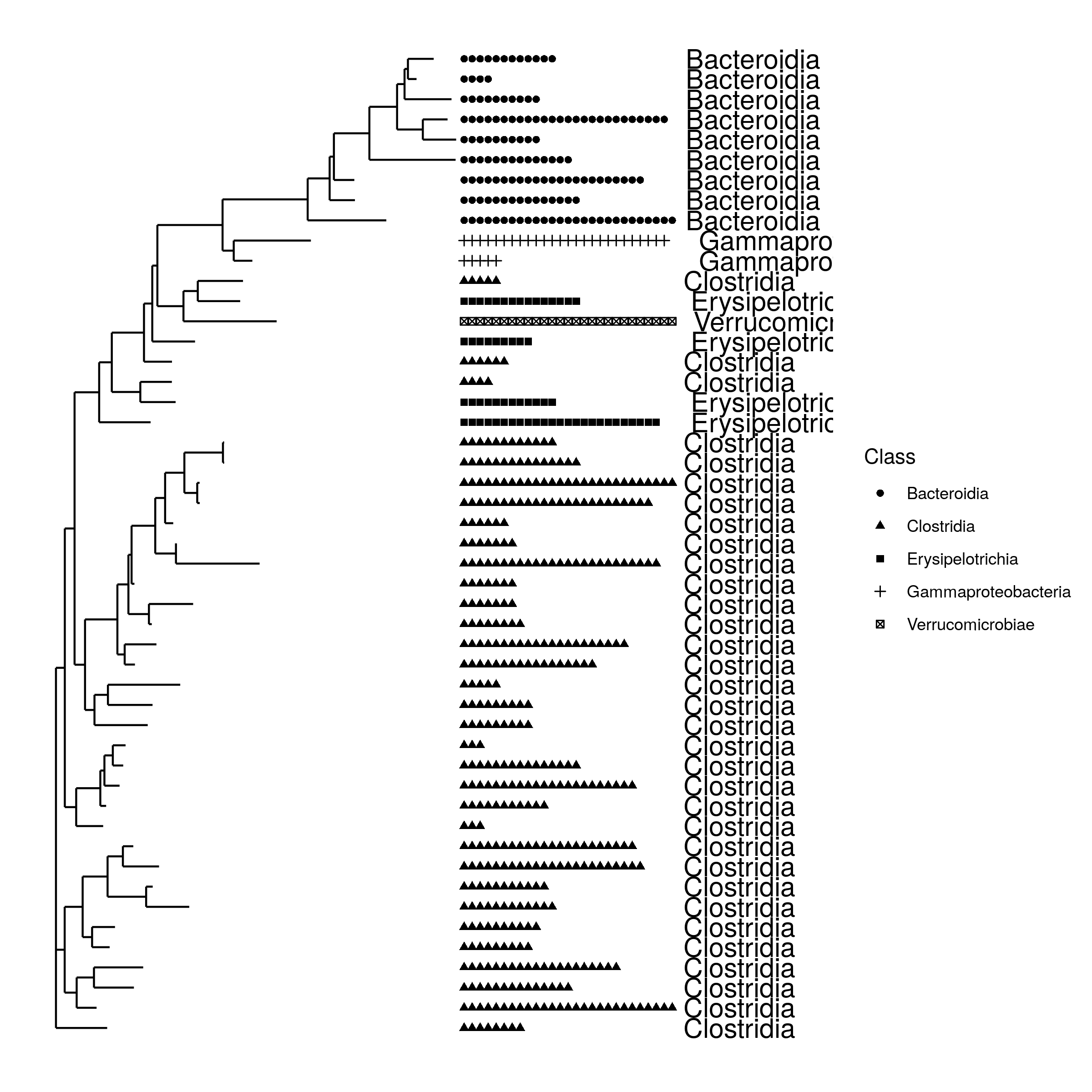
- trees
- scatter plots
- bar charts
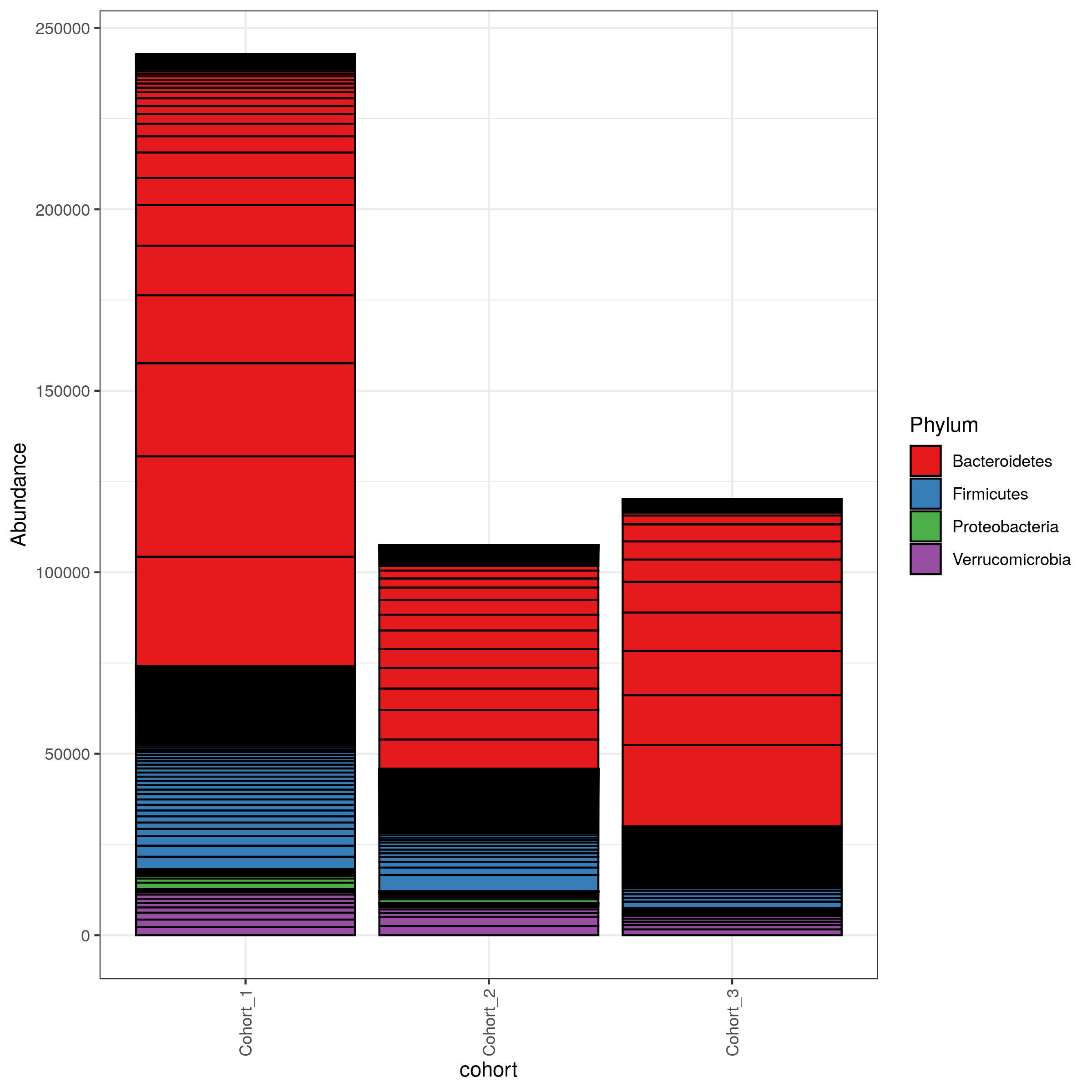
The tools is available on usegalaxy.eu (https://usegalaxy.eu/?tool_id=interactive_tool_phyloseq\&version=latest) and was integrated into a dada2 based GTN tutorial by Bérénice Batut.
Data upload from Galaxy
Although the IT worked with the original shiny app. The shiny-phyloseq app did not allow to upload amplicon data using command line arguments, which makes it difficult to allow for data input from Galaxy directly (downloading data from the Galaxy History and uploading to the IT via the web frontend would work though). Therefore, the tool was forked and modified to allow for additional data input. This functional adaptation of shiny-apps to allow for non-web based data upload is the only requirement to make any shiny app compatible with Galaxy based data upload.
The changes made to the original app can be found in this git diff.
Demo of the IT
Outlook
To improve the usability and Galaxy-interaction of the phyloseq-shiny app we are working on the possibility to export data (figures, modified phyloseq objects) to the Galaxy history, either by collecting the data as outputs of the IT or by using utility functions such as put / get, that are used in other ITs like RStudio.