InteractoMIX
Launch Galaxy| Public Server: | InteroMIX server (IT) |
|---|---|
| Scope: | Domain Specific |
| Summary: | A one-stop resource offering a wide range of computational analyses from proteome-wide interactomes to structure of protein complexes |
Comments
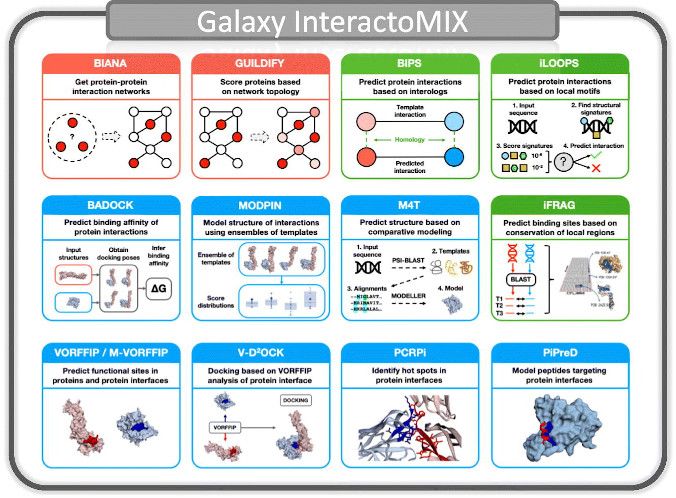
- InteractoMIX features a set of tools designed to address two major aspects in the study of protein-protein interactions (PPIs)
- i. What proteins interact. That is, the study of PPIs as a network of interactions including integration of interactomics data, characterization, prediction and application to drug discovery.
- ii. How proteins interact. That is, the structural details of PPIs, including characterization of protein interfaces, prediction of sites, docking and peptide design.
User Support
Quotas
- Anonymous access is supported. You can also create a login.
- Unregistered user : you can use the available tools. Allocated storage space is limited to 20 Mb
- Registered user : you can use the available workflows and save histories, and work with more data
Citations
- Mirela-Bota, P., Aguirre-Plans, J., Meseguer, A., Galletti, C., Segura, J., Planas-Iglesias, J., Garcia-Garcia, J., Guney, E., Oliva, B., & Fernandez-Fuentes, N. (2020). Galaxy InteractoMIX: An Integrated Computational Platform for the Study of Protein–Protein Interaction Data. Journal of Molecular Biology, 166656. DOI: 10.1016/j.jmb.2020.09.015
- InteractoMIX tagged publications in the Galaxy Publication Library
Sponsors
- InteractoMIX is the result of the combined efforts of Structural Bioinformatics and Bioinsilico labs