Multi-tenant, multi-project GVL with Galaxy, Jupyter, Terminal, and RStudio
Complete analysis on one platform, including training and collaboration
We are happy to announce the latest release of the Genomics Virtual Lab (GVL), beta3, that brings two new major features:
- Isolated projects, and
- Multiple applications.
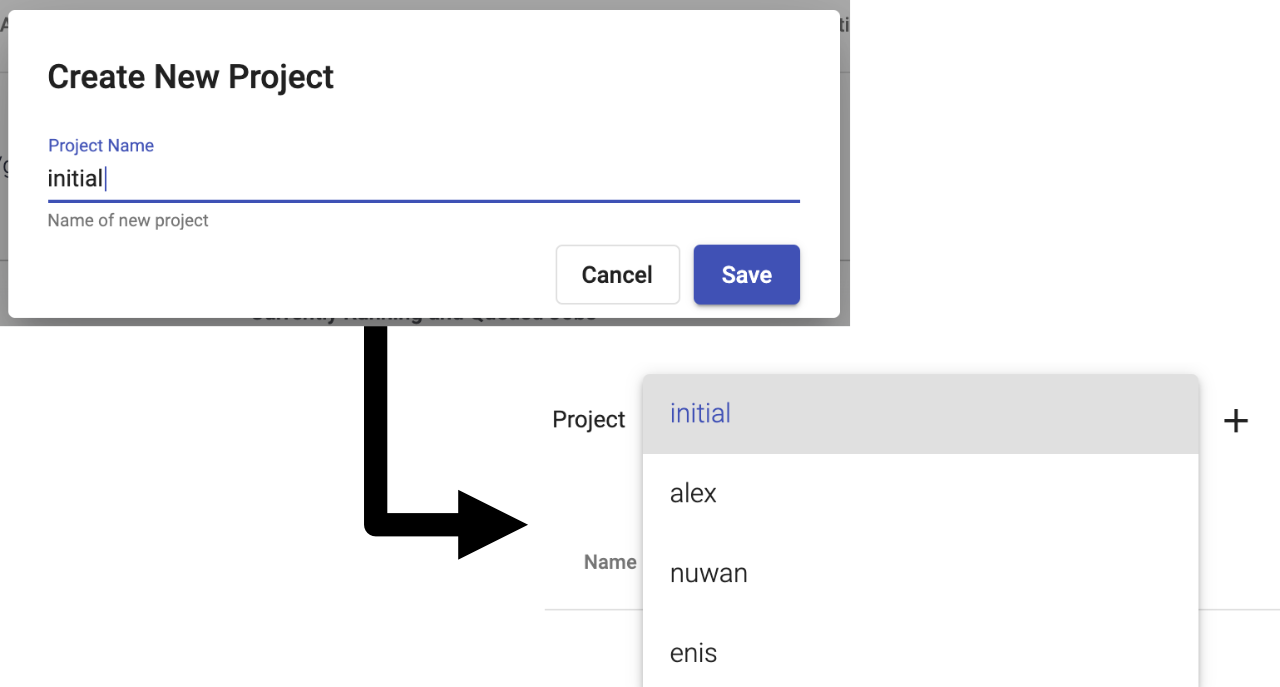
GVL projects
Traditionally, the GVL has provided a mechanism for users to obtain ready-access to a production-grade Galaxy installation on the cloud in a self-serve model. In that model, users would launch their own instance, first needing to obtain access to a cloud provider, procure cloud credentials, navigate the launch process, and then maintain the launched instance in the long term. The GVL 5.0 release aims to relieve the user from that burden by providing a platform on which managed GVL deployments can be made available by groups and institutions. With a managed GVL, researchers simply start a Galaxy instance, as opposed to launching it. The difference between start and launch is that the managed GVL can provide the cloud resources and the management software as already running services where users simply claim access to a new Galaxy instance, running in an isolated containerized environment, and do not need to provide their own resources or cloud credentials. In many ways it is similar to how *usegalaxy.** servers provide access to a ready-to-use Galaxy. With the GVL, the goal is to provide a similar service of easy-to-access Galaxy but for private Galaxy instances.
As a step in this direction, the beta3 release of the GVL 5.0 series introduces the notion of projects. A project is a virtual separation of the environment in which Galaxy runs. Projects allow multiple Galaxy instances to run simultaneously on the same underlying resources while each instance is completely separate. With this capability, individual users or different groups can obtain a Galaxy instance by simply requesting a new GVL project, without providing cloud credentials or managing the cloud infrastructure. A GVL administrator creates a new project and grants the user access. Each Galaxy instance has its own database, data, and users, separated at the infrastructure level. This provides isolation and flexibility where different Galaxy instances can run differentiated toolsets, be supported by different cloud allocations, or be made available to targeted users.
While a managed deployment of the GVL that anyone can access and request their Galaxy instance is still an endeavor for the future, the GVL projects are now available on latest GVL instances launched via CloudLaunch. In turn, the project feature caters to the classroom or workshop training use cases where individual participants can have access to their own Galaxy instance, regardless if they are using Galaxy or administering Galaxy.
Support for multiple applications
In addition to the production installation of Galaxy, the GVL now offers users access to Jupyter, RStudio, and a web-based Terminal within any GVL project. A GVL administrator has an option to add applications at-will via Helm Charts. This multi-application environment significantly expands the capabilities for data analysis with the GVL. Researchers can seamlessly switch between these applications and work in an environment most suitable for the task at hand: interactive Python scripts are within reach in a Jupyter notebook allowing users to mix code, figures, and text; some 1,500 Bioconductor packages are available for installation in RStudio; and one-off scripts can be developed in the Terminal. Meanwhile, Galaxy remains as a graphical interface to workflows and well-known tools with extensive sharing capabilities. As a user, you have unrestricted access to those applications and can install any additional packages within those applications that are needed for the analysis.
Having access to multiple applications on the same platform needs to be complemented with the ability to easily exchange data between those applications. In turn, each GVL project comes with a project-scoped data folder that is available in any of the available applications, mounted under /gvl/projects/current/. For example, a file formatted in the Terminal is immediately visible in RStudio and can be readily visualized and ingested for analyses. Similarly, the shared folder can be made available in a Galaxy data library, although not automatically yet, and subsequently imported into a History for analysis. The directory being consistent across apps and projects, ensures the portability of scripts expecting files at that path.
To further facilitate classroom or workshop usage scenarios, each GVL comes with a folder that is shared across all projects, mounted at /gvl/public. Any file placed in this directory will be simultaneously available in any GVL project. This folder provides a medium to share common inputs and training materials, or final results as outcomes from a lecture.
Use these GVL features
To get started with these features, launch the latest release of the GVL on your choice of cloud provider using CloudLaunch. After the server starts up (~12 minutes on AWS), Galaxy will automatically start and be available at the link provided. To start Jupyter, RStudio, or Terminal, click the Add Application button on the CloudMan dashboard and select which application you’d like to add. In a few moments, a link to the selected application will become available and the application will be ready for use about a minute later.
As with the recent GVL releases (Feb, Apr), we are still in beta but we are approaching the end of adding new features. We will then transition to improving robustness of the applications as well as write documentation. In the meantime, please take the GVL for a test drive and let us know how it feels. If you have a good use case that makes use of the multiple applications now available, please reach out to the authors. We’d love to work with you to support the use case, test the platform, and ideally write a paper describing the new capabilities.