Whether you are analysing microbiome samples or bacterial isolates, long reads or short, shotgun or 16S, genomics, transcriptomics, proteomics or metabolomics, multiomics or integrative analysis this is the place to be!
microGalaxy is a community of practice in the Galaxy community for anything regarding microbial data analysis in Galaxy!
What is available?
Tools
Many tools for microbial data analysis are freely available in the ToolShed which can be installed on any Galaxy server. To keep an overview, the microGalaxy community of practice maintains a curated list of the 200+ tools available.
If tools are missing or information is not up-to-date in the list, please help us! Contact Saskia, Paul or Bérénice about it.
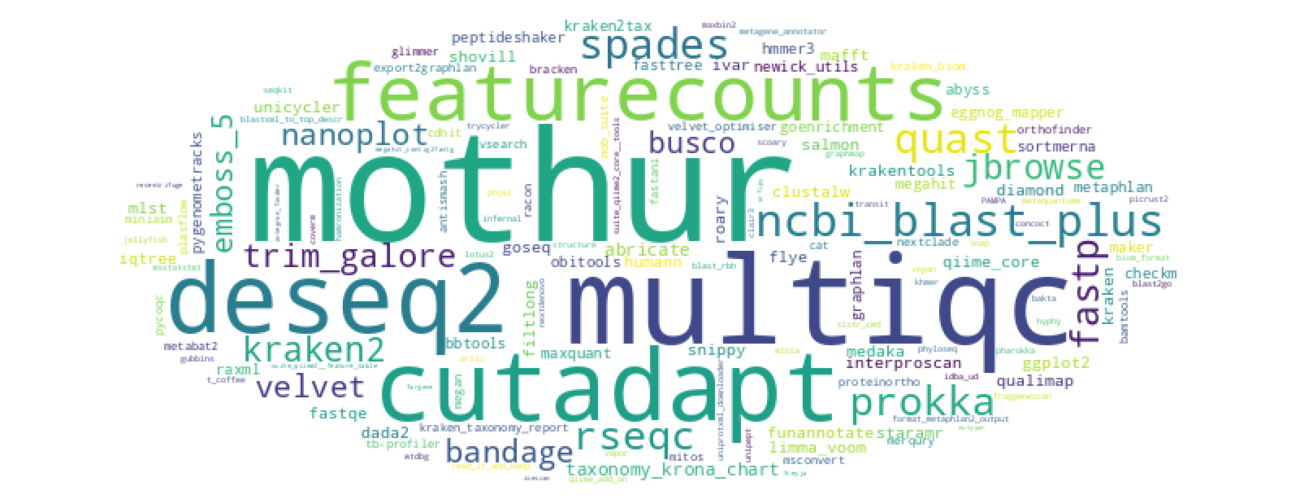
The wordcloud is based on the number of users that used the shown tools since beginning of 2022 until Sept. 2023. The usage numbers were queried from useglaxy.eu and the tools comprise all dedicated tools selected for this community.
Workflows and tutorials
Several curated Galaxy workflows are publicly available for different kinds of microbial data analysis. Many of these are accompanied by comprehensive GTN Tutorials that will guide you through the analysis step by step.
The lists below are far from being exhaustive and up-to-date. Something missing? Feel free to update the page or reach to us
Microbiome
| Target | Technique | Sequencing | Analysis | GTN Tutorial | Workflow | Notes |
|---|---|---|---|---|---|---|
| Bacteria | 16S | Short reads | Taxonomic Profiling | mothur SOP | ||
| Bacteria | 16S | Long reads | Taxonomic Profiling | |||
| Bacteria, Fungi | SSU, LSU and ITS | Short reads | Taxonomic Profiling and Visualization | WIP | WIP | Amplicon part of MGnify v5 |
| All | Shotgun metagenomics | Short reads | Taxonomic Profiling, Functional Analysis | ASaiM Docs | ||
| Plasmid | Shotgun metagenomics | Long reads | AMR detection | |||
| Beer microbiome | Shotgun metagenomics | Long reads | Taxonomic profiling | |||
| All | Shotgun metatranscriptomics | Short reads | Functional Analysis | ASaiM-MT | ||
| Pathogens | Shotgun metagenomics | Long reads | Taxonomic Profiling, Gene Identification, SNP identification, Pathogen Tracking | |||
| All | Shotgun metagenomics | Short reads | Assembly | |||
| All | Shotgun metagenomics | Short reads | Taxonomic Profiling and Visualization | |||
| Bacteria | Shotgun metagenomics, metatranscriptomics, metaproteomics | Short reads | MAGs builing, Functional analysis, Multiomics analysis | NA | ViMO (Visualizer for Meta-Omics) |
Bacterial Isolates
| Target | Sequencing | Analysis | GTN Tutorial | Workflow | Notes |
|---|---|---|---|---|---|
| M. tuberculosis | Short reads | Variant Detection | |||
| M. tuberculosis | Short reads | Transmission of Mtb | |||
| M. tuberculosis | Short reads | Phylogenetics applied to Mtb evolution & epidemiology | |||
| Plasmids | Long reads | AMR detection |
Want to include your workflow here? Let us know!
A dedicated interface
A dedicated interface with tools, workflows for microbial data analyses can be accessed via microgalaxy.usegalaxy.eu
Training material and events
The Galaxy community organizes regularly training events. You can check the event pages here to get the last ones.
We would like to highlight few events related to microbial data analysis:
GTN Smörgåsbord 2023: a free, online, self-paced, choose-your-own-adventure style, 1-week event in May 2023
There are several tracks that might be interesting for you:
How is Galaxy used for microbial data analysis?
Projects / Showcases
Below is a list of projects involving members of this community:
| Project | Description | Techniques | Sequencing | Analyses | People involved | Funding | Status |
|---|---|---|---|---|---|---|---|
| IRIDA | The Integrated Rapid Infectious Disease Analysis | Aaron Petkau | |||||
| Foodborne pathogen detection | Metagenomics | Long reads | Bérénice Batut, Engy Nasr | EOSC-Life for 2022 | Ongoing | ||
| BeerDEcoded | General public education project | Metagenomics | Long reads | Bérénice Batut, Street Science Community | Mozilla, de.NBI | Ongoing | |
| DNAnalyzer | Online & interactive game on DNA data analysis | Metagenomics | Long reads | Bérénice Batut, Street Science Community | University of Freiburg | Ongoing | |
| Cloud data | Metagenomics, metatranscriptomics | Short reads | Taxonomy profiling, Functional profiling, Assembly, MAGs builiding | Bérénice Batut, Engy Nasr | Ongoing | ||
| ASaiM | Metagenomics, metatranscriptomics | Short reads | Taxonomy profiling, Functional profiling | Bérénice Batut, Saskia Hilteman, Pratik Jagtap, Subina Mehta, etc | Finished? | ||
| Mycobacterium tuberculosis NGS made easy: data | Series of modules containing recorded videos by experts and tutorials | Illumina (wanting to expand to nanopore) | Mapping & Variant calling, Molecular epidemiology of TB using NGS, Phylogenetics for studying TB evolution and epidemiology | Daniela Brites, Peter van Heusden, Galo A. Goig, Christoph Stritt | Initially funded trough TB projects of capacity building; currently no funding | Ready to be used but also working on improvements |
Partners
| Partners | Description | People involved |
|---|---|---|
| Seq4AMR | JPIAMR Network for Integrating Microbial Sequencing and Platforms for Antimicrobial Resistance | Saskia Hilteman |
| ELIXIR Emerging Microbiome Community | Bérénice Batut | |
| µbioinfo Slack |
microGalaxy Community

microGalaxy is a community of practice to
- Develop and Sustain microbial data analysis in Galaxy
- Implement standardised "best practices"
- Expand documentation and training
- Coordinate efforts to avoid redundancy in tools, workflows and training
- Share the amazing things we are all doing together and outside Galaxy
- Support each other
Meetings
The microGalaxy community usually meets 4 times per year (including 1 BoF at GCC and other community events) for a 2-hour meeting. The meeting time alternates between 5-7pm Central European time and 9-11am Central European time in order to support members leaving in different timezones. The main aims of the meetings are to
- Get news and updates from the community
- Have 1 or more "short" talks about a project or a topic of interest
- Discuss and coordinate our efforts.
Details about the next events and minutes of previous meetings can be found here.
Working groups
2 working groups were started within the community to work together towards the goals of the commmunity:
- A Tool working groups with the aims to
- Create and maintain a curated up-to-date list of the available tools in Galaxy ecosystem for microbial data analysis
- Create a wishlist of tools to integrate in Galaxy
- Assess the needs from users for tools in Galaxy
- Organise hackathons to add tools in connection with the global community
- A Paper(s) working groups with the aims to
- Advertise about microbial data analysis in Galaxy
- Investigate on the uses of Galaxy for microbial data analysis
- Aggregate information about available resources and infrastructure within Galaxy community
- Reflect on a roadmap for the community
Join us
Anybody interested in microbial data analysis in the Galaxy is welcome to join microGalaxy! Everybody is Welcome!
- Join our quarterly meetings!
- Join our working groups (see above)
- Join the Gitter Chat (also available via Matrix)
- Join the mailing list microgalaxy@lists.galaxyproject.org
- Subscribe to the calendar