May 2018 Galaxy News
Welcome to the May 2018 Galactic News, a summary of what is going on in the Galaxy community. If you have anything to add to next month's newsletter, then please send it to outreach@galaxyproject.org.
GCCBOSC 2018
GCCBOSC 2018 will be held 25-30 June in Portland, Oregon, United States. This brings the 2018 Galaxy Community Conference and the Bioinformatics Open Source Conference together into a unified week-long event. If you work in open source life science or data-intensive biomedical research, then there is no better place than GCCBOSC 2018 to present your work and to learn from others.
GCCBOSC starts with two days of training with a wide range of topics nominated and selected by our communities. Training is followed by a two day meeting, with joint and parallel tracks, featuring oral presentations, posters, demos, lightning talks, birds-of-a-feather and invited keynotes. The week finishes with CollaborationFest Core and Encore, two or four days of collaborative work on code, documentation, training and challenging data analysis problems.
GCC2018 schedule posted
The schedule of accepted long talks for GCC2018 is now available online. Sixteen topics have been scheduled spanning the full Galactic spectrum from deployment details to novel applications of Galaxy and everything in between.
BOSC talks and GCC lightning talks will be added in the next week or so.
Early registration ends May 11
Did you catch that? Early registration ends May 11! So c'mon, register already:
- Register
- Book conference housing
- Childcare. Space in childcare is very limited.
If you register after May 11, you will have less money to fund reagents, storage, compute timepost-docs, field work, 3D printers, VR systems...
BOSC deadline for late-breaking lightning talks, demos and posters is May 17
- Submit your abstract here.
GCCBOSC 2018 Sponsors
We are happy to have sponsors contributing to the success of GCCBOSC 2018. This month we are highlighting Advanced HPC and GigaScience.
GigaScience and Oxford University Press
GigaScience is an online open access, open data, open peer-review journal published by Oxford University Press and BGI. The journal offers ‘big data’ research from the life and biomedical sciences, and on top of 'Omics research includes the growing range of work that uses difficult-to-access large-scale data, such as imaging, neuroscience, ecology, systems biology, and other new types of shareable data. GigaScience is unique in the publishing industry as it publishes all research objects (data, software tools, source code, workflows, containers and other elements related to the work underpinning the findings in the article). Promoting Open Science, all published software needs to be under an OSI-license, all supporting data must be available and open, and all peer review is carried out transparently. Presenting workflows via our GigaGalaxy.net server, novel work presented at the meeting utilising Galaxy is eligible to a 15% APC if it is submitted to our Galaxy series.
Advanced HPC
Life sciences is an ideal domain to take full advantage of all the things that HPC technology can do. Universities, research organizations, and biotechnology companies are embracing predictive analytics, machine learning and artificial intelligence to make major gains in fighting cancer, personalizing medicine and regenerating organs. The technology is paying huge dividends. For example, research institutions that are using Illumina’s® DNA sequence and array-based technologies for genotyping and epigenetic applications are leveraging our compute and parallel solutions to get better and faster results.
Advanced HPC's experts offer many advantages, including extensive consultation and support. As HPC software specialists, our engineers can customize complex solutions for you more quickly and efficiently than our competitors.
Advanced HPC is pleased to be the first ever GCCBOSC Icebreaker Sponsor.
[
 ](https://depot.galaxyproject.org/hub/attachments/documents/gccbosc2018_sponsorship_summary_20170906.pdf) [
](https://depot.galaxyproject.org/hub/attachments/documents/gccbosc2018_sponsorship_summary_20170906.pdf) [ ](https://depot.galaxyproject.org/hub/attachments/documents/gccbosc2018_sponsorship_prospectus_20170906.pdf)
](https://depot.galaxyproject.org/hub/attachments/documents/gccbosc2018_sponsorship_prospectus_20170906.pdf) Call for sponsors!
Sponsors are a key part of GCCBOSC 2018. Is your organization interested in playing a prominent role in the first joint gathering of the Galaxy and BOSC communities? Then become a GCCBOSC 2018 sponsor and raise your organization's visibility in these active and engaged communities.
ELIXIR Workshop for Galaxy training material and skills improvement
The ELIXIR Workshop for Galaxy training material and skills improvement will be held 21-23 May at the Earlham Institute in Norwich, UK.
Education and training is an integral part of the Galaxy community. The Galaxy Training Network (GTN) has been working for several years with GOBLET and the ELIXIR Training Platform to create material and deliver training for scientists, developers and system administrators. A selection of 72 tutorials, developed by more than 60 contributors, is already available. In this workshop, we intend to improve the participants’ understanding of learning principles, training techniques and best practices for materials preparation. We will then work together to expand the existing collection of Galaxy training materials by covering more topics relevant to the ELIXIR use cases, add missing annotation and metadata and make the materials easily accessible.
The workshop will start with a 1-day "Train the Trainer" course, similar to the Carpentries' Instructor Training and the ELIXIR-EXCELERATE Train the Trainer. Here Galaxy trainers will develop an insight into different learning styles, understand what makes a good trainer, and learn new approaches to training from experienced trainers. This course will help them to better contribute and shape the materials during the hackathon.
Galaxy Africa Report

The beginning of April saw a flocking of a large number of individuals interested in all things Galaxy to Cape Town! This exciting destination was home for the first ever Galaxy Africa event that aimed to inform, educate, and bring together bioinformaticians, biologists, and computer scientists from around the African continent. Organized by SANBI and held at the University of the Western Cape, under the close eye of Peter van Heusden, nearly 50 people participated in a packed schedule where everything from an introduction to Galaxy, training, analysis, experiences, and system management were covered. There were a number of BoF sessions as well to have open discussions and start future conversations. The week ended with a Data Carpentry R workshop event.
Even though the event ended, it planted a seed for a new Galaxy community that spans the African continent: emails were matched with faces, mailing lists were formed, meals shared, and lessons passed on. Thank you Galaxy Africa!
Upcoming events
These and other Galaxy related events coming up in the next few months:
Publications
132 new publications referencing, using, extending, and implementing Galaxy were added to the Galaxy Publication Library in April.
Highlighted Publications
April added three Galactic and Stellar publications:
- IRProfiler – a software toolbox for high throughput immune receptor profiling, Christos Maramis, Athanasios Gkoufas, Anna Vardi, Evangelia Stalika, Kostas Stamatopoulos, Anastasia Hatzidimitriou, Nicos Maglaveras and Ioanna Chouvarda. BMC Bioinformatics (2018) 19:144, doi:10.1186/s12859-018-2144-z
- SECIMTools: a suite of metabolomics data analysis tools, Alexander S. Kirpich, Miguel Ibarra, Oleksandr Moskalenko, Justin M. Fear, Joseph Gerken, Xinlei Mi, Ali Ashrafi, Alison M. Morse and Lauren M. McIntyre. BMC Bioinformatics (2018) 19:151, doi:10.1186/s12859-018-2134-1
- Notos - a galaxy tool to analyze CpN observed expected ratios for inferring DNA methylation types, Ingo Bulla, Benoît Aliaga, Virginia Lacal, Jan Bulla, Christoph Grunau and Cristian Chaparro, BMC Bioinformatics (2018) 19:105, doi:10.1186/s12859-018-2115-4
Publication Topics
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 79 | +Methods | 29 | +UsePublic | 17 | +Workbench | 16 | +UseMain | |||
| 11 | +Reproducibility | 9 | +RefPublic | 7 | +UseLocal | 5 | +Other | |||
| 5 | +IsGalaxy | 5 | +Tools | 4 | +Cloud | 2 | +Shared | |||
| 1 | +Unknown |
Who's Hiring

The Galaxy is expanding! Please help it grow.
- Software Developer and Bioinformaticians, Quadram Institute, Norwich , United Kingdom
- Ingénieur en Bioinformatique NGS, Institut de Biologie Intégrative de la Cellule, Orsay, France
- Scientist (f/m), NGS bioinformatics core facility, Helmholtz Zentrum München, Germany.
- Software developer (f/m) position at the NGS bioinformatics core facility, Helmholtz Zentrum München, Germany.
- Ingenieur Bioinformatique Evolutive LIRMM, Montpellier, France
- Post-Doc / IR analyse de données miRNA, Unité de Nutrition Humaine, Clermont-Fd, France
- Freiburg Galaxy Team has open positions, Freiburg, Germany
- Development of innovative algorithms to process real-time mass spectrometry data for the clinical analysis of exhaled breath, CEA, Saclay, France
- The Blankenberg Lab in the Genomic Medicine Institute at the Cleveland Clinic Lerner Research Institute is hiring postdocs.
- Galaxy Project is hiring software engineers and postdocs at Johns Hopkins, Baltimore, Maryland, United States
Have a Galaxy-related opening? Send it to outreach@galaxyproject.org and we'll put it in the Galaxy News feed and include it in next month's update.
Public Galaxy Server News
There are over 100 publicly accessible Galaxy servers and six semi-public Galaxy services. Another public server was added to the directory in April:
IRProfiler
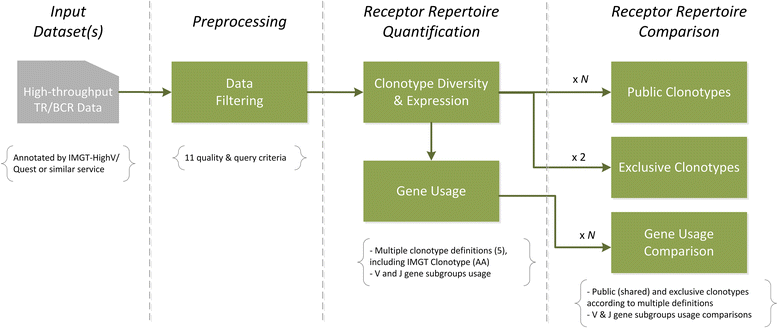
Immune Repertoire Profiler (IRProfiler) is a novel software pipeline that delivers a number of core receptor repertoire quantification and comparison functionalities on high-throughput TR and BcR sequencing data. IRProfiler can be used anonymously or through an account.
This server is made available as supplementary material for the article "IRProfiler - A Software Toolbox for High Throughput Immune Receptor Profiling", authored by C. Maramis et al. in BMC Bioinformatics. A Docker image of the IRProfiler Galaxy toolbox and wrapped IRProfiler tools are also available.
IRProfiler is supported by the Lab of Computing, Medical Informatics & Biomedical-Imaging Technologies, Department of Medicine, Aristotle University of Thessaloniki, in Thessaloniki, Greece; and the Institute of Applied Biosiences, Centre for Research & Technology Hellas, in Thermi, Greece
Public Servers in Publications
We tag papers that use, mention, implement or extend public Galaxy Servers. Here are the counts for March's publications.
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 13 | >Huttenhower | 8 | >RepeatExplorer | 4 | >Workflow4Metabolomics | 2 | >RiboGalaxy | |||
| 2 | >PRABI | 1 | >NCGAS | 1 | >Langille | 1 | >EpiToolKit | |||
| 1 | >Pasteur | 1 | >NGS-QC | 1 | [>Globus Genomics](https://www.zotero.org/groups/1732893/galaxy/tags/>Globus Genomics) | 1 | >PlantGenie | |||
| 1 | >ARGalaxy | 1 | >Osiris | 1 | >IRProfiler | 1 | >Cistrome | |||
| 1 | >Jetstream | 1 | >Alveo | 1 | [>RNA Workbench](https://www.zotero.org/groups/1732893/galaxy/tags/>RNA Workbench) |
Tools
ToolShed Contributions
 ](http://toolshed.g2.bx.psu.edu/)
](http://toolshed.g2.bx.psu.edu/)Tool Shed contributions in April.
Releases
BioBlend 0.11.0
BioBlend is a Python library for interacting with CloudMan and Galaxy‘s API. BioBlend makes it possible to script and automate the process of cloud infrastructure provisioning and scaling via CloudMan, and running of analyses via Galaxy.
See the release notes for what's new in release 0.11.0.
Starforge 0.3.6
Starforge is a collection of scripts that supports the building of components for Galaxy. Specifically, with Starforge you can:
- Build Galaxy Tool Shed dependencies
- Build Python Wheels (e.g. for the Galaxy Wheels Server)
- Rebuild Debian or Ubuntu source packages (for modifications)
These things will be built in Docker. Additionally, wheels can be built in QEMU/KVM virtualized systems. Documentation can be found at starforge.readthedocs.org.
The release features Python 3-compatible setup.py wrapping and a fix for pip 10.
Earlier Releases
Other packages released in the prior 4 months.
18.01 Galaxy Release
The 18.01 release of Galaxy is out. Thanks to the Galaxy Committers and you, our community!
Highlights:
Performance and User Experience Improvements We made Galaxy more lively and responsive. Homepage, published workflows, published/saved histories, and data libraries should all load much faster now. Importing data from FTP will also take less of your time.
Web Server and Configuration The default web server used by Galaxy has changed from Paste to uWSGI and the default configuration file for Galaxy is now config/galaxy.yml instead of config/galaxy.ini. To minimize the impact of this change on existing Galaxy instances, if a Galaxy has a galaxy.ini file configured, it will continue to use Paste by default unless additional steps are taken by the administrator
Dataset Collection Usability This release has significantly improved the usability of Galaxy dataset collections. Dozens of improvements to collections have been made, some of the key highlights include:
- Data library folders can now be sent to histories as a dataset collection.
- Failed dataset collection elements can now be fixed using job re-running
- Collections now appear with state and progress bars in the history panel and contained datasets are hidden by default
- We added intuitive workflow post job actions for dataset collections
- The web interface now supports collections with arbitrary nesting and size
- More robust nametag discovery and propagation when using collections
Client Architecture The architecture for the client code that powers the Galaxy user interface has been significantly overhauled. The code base has been converted to ES6, Yarn now powers the build and dependency management of the code, Prettier is now used to ensure consistent code formatting, and the VueJS framework has been integrated.
New BAM datatypes Previously Galaxy only supported coordinate sorted BAM files by default (the bam datatype). In addition, this release of Galaxy now supports three new types of BAM:
- qname_sorted.bam, that ensures that the file is queryname sorted (e.g. SO:queryname);
- qname_input_sorted.bam, that can be used to describe the output of aligners which generally keep mate pairs adjacent
- unsorted.bam, that makes no assumptions about the sort order of the file.
Experimental Job Caching Galaxy can now be configured to allow users the option of skipping duplicated jobs if one with identical parameters has been previously executed and simply reuse the previously generated outputs.
See the full release notes for more.
Thanks for using Galaxy!
Galaxy Docker Image 18.01
The Galaxy Docker project has a matching release, for Galaxy 18.01. The release features the 18.01 enhancement and removes nodejs-legacy and npm from the Dockerfile and installs the latest version from ansible-extras.
blend4j 0.2.0
blend4j is a partial reimplementation of the Python library bioblend for the JVM. bioblend for Python is a library for scripting interactions with Galaxy.
- Update to galaxy-bootstrap 0.7.0.
- Added support for accessing Galaxy Tool Data tables. Thanks to Dan Fornika.
- blend4j now requires Java 1.8+ to run.
galaxy-lib 18.5.3 - 18.5.7
galaxy-lib is a subset of the Galaxy core code base designed to be used as a library. This subset has minimal dependencies and should be Python 3 compatible. It's available from GitHub and PyPi.
Planemo 0.48.0
Planemo is a set of command-line utilities to assist in building tools for the Galaxy project. These releases included numerous fixes and enhancements.
See GitHub for details.
Pulsar 0.8.1-3
Pulsar updates were released in February. Pulsar is a Python server application that allows a Galaxy server to run jobs on remote systems (including Windows) without requiring a shared mounted file systems. Unlike traditional Galaxy job runners - input files, scripts, and config files may be transferred to the remote system, the job is executed, and the results are transferred back to the Galaxy server - eliminating the need for a shared file system.
And the rest ...
Other Galaxy packages that haven't had a release in the past four months can be found on GitHub.