This content has a new home at /learn.
Uploading data into Galaxy
Uploading data into Galaxy can be done in number of ways summarized in the following chart:
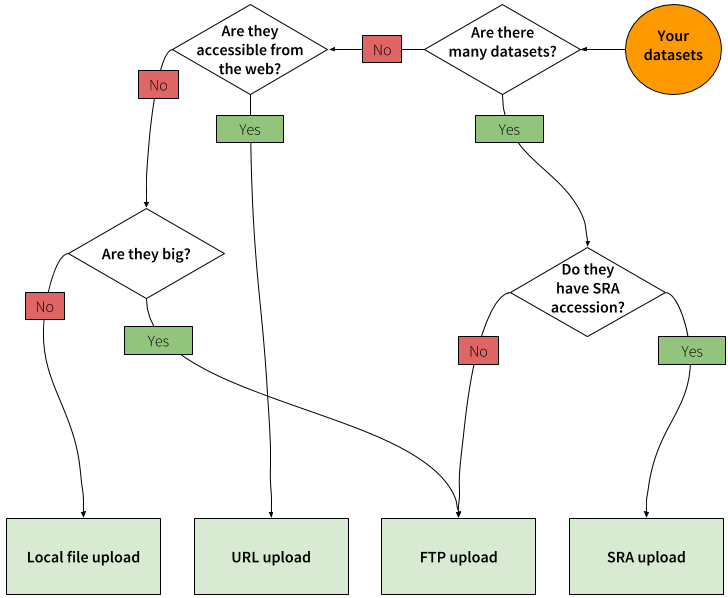 |
| Figure 1. Deciding which upload approach to use depends on the size and number of datasets, whether they are web accessible, or if they have been deposited to the Short Read Archive (SRA). Many = more than 10. Big = over 100 Mb. |
The general "best practice approach" is this:
- if you have just a few small (< 100 MB) datasets stored on your computer → use the Local file upload
- if you have large files (> 100 MB) → use FTP upload
- if you have many files (> 10) → use FTP upload
- if datasets you are uploading have been deposited to SRA → use SRA upload
Local file upload
Uploading local files directly into Galaxy works well for a small number (say, a dozen) or relatively small (tens of MB) datasets. The following screencast shows how this works:
FTP upload
FTP stands for file transfer protocol. This is the best way to upload large files (or large number of files) into Galaxy:
URL upload
In some cases you may need to upload public datasets from the Internet. These datasets will have web addresses (also called Uniform Resource Locators or URLs). A web address of a dataset can be pasted directly into the upload tool interface:
SRA upload
Finally, if the data you are uploading has been deposited to the Short Read Archive (SRA) at NCBI use the following approach:
Note: SRA approach downloads individual datasets into collections. Galaxy uses collections to combine similar datasets in order to streamline interface and minimize the amount of clicking you need to do. Please, take a look a collections tutorial to understand this useful concept.
To GZIP or not to GZIP
Compressing can be a highly efficient way to store many types of biological data. For example fastq datasets (a typical representation for sequencing reads) can be compressed to approx. 33% of their original size with gzip utility. So if the data you are uploading is already compressed (e.g., has .gz or .bz2 file extensions) keep it this way! Upon upload simply tell Galaxy (as shown in FTP upload video) that file(s) is(are) compressed (i.e., of fastqsanger.gz type if this is appropriate).