April 2013 Galaxy Update
Welcome to the April 2013 Galaxy Update, a monthly summary of what is going on in the Galaxy community. Galaxy Updates complement the Galaxy Development News Briefs which accompany new Galaxy releases and focus on Galaxy code updates.
New Public Galaxy Servers
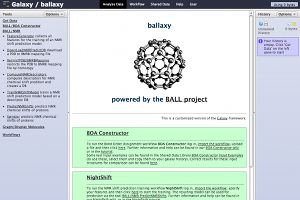
ballaxy hosts the BALL (Biochemical Algorithms Library) Project tools, i.e. computer aided drug design and molecular modelling based on protein and ligand structure data. It is tailored to handle structural molecular data (pdb, mol, mol2, hin, xyz, smiles) and to offer tools for modelling tasks like chemical shift prediction "NightShift" or optimal bond order assignment of ligands "BOA Constructor". A login is required and everybody can create a login, but there is no guarantee how long any data will be preserved. ballaxy is supported by the groups of Hans-Peter Lenhof (Saarland University, Saarbrücken, Germany), Oliver Kohlbacher (University of Tübingen, Germany), and Andreas Hildebrandt (University of Mainz, Germany).
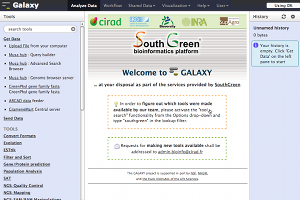
South Green Galaxy is a part of the South Green Bioinformatics Platform (SGBP), "a bioinformatics platform applied to the genomic resource analysis of southern and Mediterranean plants." It supports many custom tools and data sources relevant to plants. It also has a 10 MB storage quota for anonymous users. Account creation is restricted to those with an active collaboration and that work on southern and Mediterranean plants. South Green Galaxy is sponsored by CIRAD, IRD (Institut de recherche pour le développement), Bioversity International, INRA, Montpellier SupAgro
These servers join the growing list of public Galaxy servers.
New Papers
| # | Tag |
|---|---|
| 16 | methods |
| 8 | usemain |
| 7 | workbench |
| 4 | unknown |
| 3 | tools |
| 2 | isgalaxy |
| 2 | shared |
| 2 | usepublic |
| 2 | cloud |
| 1 | project |
| 1 | uselocal |
There are several things to report in the New Papers arena. First, the process we used to synchronise the Galaxy CiteULike group with it's Mendeley mirror broke, and has not been fixed yet. Mendeley dropped support for automatic synchronization with CiteULike at the end of February and so no updates have been pushed to Mendeley since then. We are looking for processes that are not too labor / manual intervention intensive. We are also reviewing our general use of CiteULike and Mendeley. If you have any suggestions, please [send them to us](mailto:outreach AT galaxyproject DOT org).
Second several new tags were added, most of them attached to papers with the methods tag, and indicating (if the paper indicates) where the analysis was run.
usemain: |
Analysis run on Main |
usepublic: |
Analysis run on other public Galaxy servers |
useprivate: |
Analysis run on private Galaxy servers |
usecloud: |
Analysis run on cloud based Galaxy servers |
cloud: |
Papers that emphasize or are about cloud deployments |
We will likely back-annotate papers with these new tags from the beginning of 2013, but not prior to that.
Finally, 32 new papers were added to the Galaxy CiteULike Group in March, including these which might be of general interest to the Galaxy community.
- "NightShift: NMR Shift Inference by General Hybrid Model Training - a Framework for NMR Chemical Shift Prediction" by Dehof et al. BMC Bioinformatics, Vol. 14, No. 1. (16 March 2013), 98
- "Computational Predictions Provide Insights into the Biology of TAL Effector Target Sites" by Grau et al. PLoS computational biology, Vol. 9, No. 3. (14 March 2013), e1002962
- "Aye-aye population genomic analyses highlight an important center of endemism in northern Madagascar," by Perry, et al. Proceedings of the National Academy of Sciences (25 March 2013)
There are now over 900 papers in the Galaxy CiteULike Group.
Who's Hiring
The Galaxy is expanding! Please help it grow.
- The Galaxy Project is hiring software engineers and post-docs! at both Emory and Penn State.
- Senior Developer, Stem Cell Bioinformatics Core : Seattle, WA, United States.
- CDD Ingénieur en Bioinformatique grand séquençage - Institut Curie
- Two openings @ LSU: Bioinformatics Support Group Leader and Post-doc/Res Assoc
- Bioinformatics Trainter/Analyst, Morgridge Institute for Research, Madison, Wisconsin, United States
Got a Galaxy-related opening? Send it to outreach@galaxyproject.org and we'll put it in the Galaxy News feed and include it in next month's update.
GCC2013
| The deadline for submitting oral presentation abstracts is 12 April. |
|---|
The 2013 Galaxy Community Conference (GCC2013) will be held 30 June through July 2 in Oslo Norway, at the University of Oslo.
GCC2013 is an opportunity to participate in two full days of presentations, discussions, poster sessions, keynotes, lightning talks and breakouts, all about high-throughput biology and the tools that support it. The conference also includes a Training Day for the second year in a row, this year with more in-depth topic coverage, more concurrent sessions, and more topics.
Registration, Talk/Poster Submission Open
Early registration and paper and poster abstract submission are open.
Register early and save up to 75% off regular registration costs. Early registration is very affordable, with combined registration (Training Day + main meeting) starting at ~ €95 for post-docs and students. Registering early also assures you a spot in the Training Day workshops you want to attend. Once a session becomes full, it will be closed to new registrations. Early registration closes 24 May.
Abstract submission for both oral presentations and posters is also open. Abstract submission for oral presentations closes 12 April, and poster abstract submission closes 3 May. Please consider presenting your work. If you are dealing with big biological data, then this meeting wants to hear about your work.
Sponsorships
There are still several sponsorships still available. Please contact the Organizers if your organization would like to help sponsor this event.
Galaxy at GMOD 2013, Biocuration 2013 and TGAC
Galaxy will have a presence at both the [GMOD 2013](http://gmod.org/wiki/April 2013 GMOD Meeting) and Biocuration 2013. Both are being held in Cambridge, UK in early April. If you are interested in either community, then it would be great to see you there.
There is also a Galaxy Workshop at The Genome Analysis Centre in nearby Norwich, UK on 11 April, the day after Biocuration ends.
Other Upcoming Events
Events
GalaxyAdmins
March 2013 Meetup
Slides and the screencast from the March GalaxyAdmins Meetup are now available [Hailiang "Leon" Mei](mailto:hailiang DOT mei AT nbic DOT nl) spoke on NBIC Galaxy deployment at SURFsara's HPC cloud. NBIC Galaxy (Andromeda) was migrated to a HPC cloud hosted by Surfsara in September 2012. Leon discussed the setup of this HPC cloud and the architecture of NBIC Galaxy and shared their experiences installing the NBIC Galaxy using the Cloudman scripts. The presentation finished with a list of issues and our possible future plans. Dan Blankenberg also discussed the new Galaxy release process, and the upcoming Data Manager enhancements.
GalaxyAdmins Future Directions
The GalaxyAdmins group is coming up on it's one year anniversary (coinciding with GCC2013) and this is a good opportunity to discuss what the future of the group should be. If you are interested, please join the discussion on this email thread.
Some starting topics for discussion are on the GalaxyAdmins Future Directions page. These include
- What should the group's goals and activities be?
- What type of leadership structure should the group have, and how should it be selected?
The discussion, however, is wide open to any topic relevant to the group. If you have any opinions or suggestions please reply to the group. Anyone with an interest in the group is encouraged to post.
Once the discussion settles, we will summarize the discussion on the wiki page and suggest an action plan for making those things happen.
Galaxy Distributions
- Job running configuration changes and refactoring of job runner plugins
- Required reset of the metadata on your installed tool shed repositories plus Changes required to your environment if you are hosting a local Tool Shed
- Basic Data Manager Functionality now present
- Tool Shed, Trackster, Cloudman, Workflows, UI, Admin, and new Pull Request updates.
- New bug fixes and improvements.
Tool Shed Contributions
- ctcf_analysis: computationally predict CTCF sites for a nucleotide sequence
- interproscan_to_excel: Convert interproscan raw or xml output to an Excel file
- snpeff_cds_report: Get transcripts from Ensembl for snpEff variants and report coding changes
- mmuff: Missense Mutation and Frameshift Finder Workflow
Other News
- "Galaxy for Core Facilities" ABRF talk slides now available
- The servers that power Main were relocated to their new room and building on March 14.
- The Galaxy Project now has a LinkedIn group. Please consider joining.
- Several items relating to Galaxy have been approved for ISMB / ECCB 2013. Will post as soon as there is a schedule.
- Documentation for dynamic job destinations
- Cloud-Based Image Analysis and Processing Toolbox