January 2018 Galaxy News
Welcome to the January 2018 Galactic News, a summary of what is going on in the Galaxy community. If you have anything to add to next month's newsletter, then please send it to outreach@galaxyproject.org.
Events
There are a plenitude of Galaxy related events coming up in the next few months:
GCCBOSC 2018 Training Nominations Extended to January 12
Nominate a training topic now.
The joint 2018 Galaxy Community and Bioinformatics Open Source Conferences start with training and the training topics are determined by you. Topic nominations have been extended January 12. Any topics of interest to these communities can be nominated.
GCCBOSC2018 will be held 25-30 June in Portland, Oregon, United States. It will feature two days of training: the second of which is multi-track and will feature content for both the BOSC and Galaxy communities.
Nominated topics can cover a wide range. For example:
- How to use open source software using just a web browser (e.g., Intro to Using Galaxy)
- How to use software packages (The Newbie's Guide to BioRuby)
- Advanced applications of software (Genome Assembly with Galaxy, or Proteomics with BioPython)
- Software installation and configuration (Tool Wrapping for Galaxy, or Using CloudLaunch)
- Open source community (Building a Curation Community using Apollo, or Training using the Galaxy Training Network)
These are only examples. If you are looking for ideas, see topics nominated for GCC2017, GCC2016, GCC2015, and the Galaxy Events page, which lists what training the community is offering outside of GCCs. Training is not limited to Galaxy-related topics. Don't be afraid to think inside the BOSC!
Nominated topics have been, and will continue to be published as they come in. Topics will be compiled into a uniform list by the GCCBOSC2018 organizers, and then voted on by the community later this month.
Galaxy (and GMOD) at Plant and Animal Genome XXVI
The 26th Plant and Animal Genome Conference will be held January 13-17, in San Diego, California. PAG is the largest ag-genomics meeting in the world and Galaxy will be there to help researchers get the most from their data. There will be (at least):
- a Galaxy Workshop with 4 talks,
- a Galaxy Community Update talk during the GMOD session
- 10 other Galaxy-related talks, and
- 12 Galaxy-related posters.
- 4 pre-conference GMOD and Galaxy events
See the Galaxy @ PAG page for details.
GMOD @ PAG

GMOD (of which Galaxy is a part) has a bunch of things going on at PAG that are of interest to the Galaxy Community:
- GMOD will have a project-wide hackathon on January 11-12.
- Meg Staton will speak about the Tripal-Galaxy bridge
- There will be a GMOD workshop on the last day of PAG.
- The Tripal Project will have a user meeting on January 11, and a hackathon on January 12. Register here.
If you are interested in supporting better integration of Galaxy with other GMOD components then please give the two hackathons a look.
Intro to Galaxy Workshop at CSUSM
If you are at Cal State San Marcos, or anywhere nearby, there will be an Introduction to Galaxy Workshop in San Marcos, California on Friday January 12. The workshop is free, but space is limited, so you are encouraged to register now.
ELIXIR Galaxy Community Kickoff and Meeting
 ](https://www.elixir-europe.org/events/galaxy-community-kickoff-meeting-and-galaxy-user-conference)
](https://www.elixir-europe.org/events/galaxy-community-kickoff-meeting-and-galaxy-user-conference) [
 ](http://www.emblaustralia.org/)
](http://www.emblaustralia.org/)
The ELIXIR Galaxy community will have a kickoff meeting 14-16th March 2018, in Freiburg/Germany. We will combine this with a Galaxy User Conference and the official launch of the usegalaxy.eu server.
This event will also be coordinated with the EMBL Australia to also launch usegalaxy.org.au at the same time.
Wednesday (the 14th) will be dedicated to the ELIXIR Galaxy communtiy, to discuss and plan our roadmap in 2018 and 2019. Thursday and Friday (15th, 16th) will be a conference where Galaxy users talk about their research and use-cases. However, a few spots we will reserved for official talks from ELIXIR.
Register now as space is limited.
2018 Big Genomics Data Skills Training course
The Jackson Lab (JAX) is pleased to open the application period for the 2018 Big Genomics Data Skills Training course, funded by the NIH BD2K initiative: Big Genomic Skills Training info and online application. The program provides training and hands-on experience working with genomic scale data and will enable participants to integrate genomic data analysis into their courses or launch new courses. Topics covered include RNAseq, Whole Exome Sequencing and variant detection, ChIPseq and network analysis. Participants will gain experience using Galaxy, R and Python.
As a grant funded activity housing is included; however you will need to cover your own travel. If you teach at an HBCU or Minority-serving institution (by Dept. of Education criteria) JAX may be able to provide a modest travel award.
The course is for faculty who primarily teach undergraduate students and will be held at JAX Genomic Medicine in Farmington CT, May 21-25.
All Upcoming events
See the Galaxy Events Google Calendar for details on other events of interest to the community.
New Galactic Blog Entries: Galaksio
 ](/news/)
](/news/) [
 ](/news/)
](/news/)
We've got two new Galactic Blog entries, both related to Galaksio:
- Galaksio: a Galaxy user interface focused on running prepared workflows, by Tomas Klingström
- Bioinformatic training with the B3Africa project, by Tomas Klingström
The Galactic Blog is a place for longer-form posts that are relevant to Galaxy. (See Galaxy News for shorter-form items.)
Publications
155 new publications referencing, using, extending, and implementing Galaxy were added to the Galaxy Publication Library in December (and over 1500 publications were added in 2017).
Highlighted Publications
The Galactic and Stellar publications added in December were:
- TOSCA-based orchestration of complex clusters at the IaaS level, M Caballer, G Donvito, G Moltó, R Rocha and M Velten. Journal of Physics: Conference Series, Volume 898, Track 6: Infrastructures
- Molecular property diagnostic suite (MPDS): Development of disease-specific open source web portals for drug discovery, S. Nagamani, A. S. Gaur, K. Tanneeru, G. Muneeswaran, S. S. Madugula, MPDS Consortium, D. Druzhilovskiy, V. V. Poroikov & G. N. Sastry. SAR and QSAR in Environmental Research, Special Issue: 9th International Symposium on Computational Methods in Toxicology and Pharmacology Integrating Internet Resources (CMTPI-2017) - Part 2. Guest Editors: A.K. Saxena and M. Saxena
- The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics, Laurel Cooper, Austin Meier, Marie-Angélique Laporte, Justin L. Elser, Chris Mungall, Brandon T. Sinn, Dario Cavaliere, Seth Carbon, Nathan A. Dunn, Barry Smith, Botong Qu, Justin Preece, Eugene Zhang, Sinisa Todorovic, Georgios Gkoutos, John H. Doonan, Dennis W. Stevenson, Elizabeth Arnaud and Pankaj Jaiswal. Nucleic Acids Research, 2017 1 doi: 10.1093/nar/gkx1152
- FROGS: Find, Rapidly, OTUs with Galaxy Solution, Frédéric Escudié, Lucas Auer, Maria Bernard, Mahendra Mariadassou, Laurent Cauquil, Katia Vidal, Sarah Maman, Guillermina Hernandez-Raquet, Sylvie Combes, Géraldine Pascal. Bioinformatics, btx791, https://doi.org/10.1093/bioinformatics/btx791
- Transcriptomic Data Analysis: RNA-Seq Analysis Using Galaxy, Chia-Lang Hsu and Chantal Hoi Yin Cheung (2018), in A Practical Guide to Cancer Systems Biology: pp. 49-62. https://doi.org/10.1142/9789813229150\_0005
Publication Topics
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 96 | +Methods | 32 | +UsePublic | 18 | +Workbench | 14 | +UseLocal | |||
| 13 | +UseMain | 11 | +RefPublic | 7 | +Tools | 6 | +Reproducibility | |||
| 5 | +Unknown | 4 | +Cloud | 3 | +Other | 2 | +Visualization | |||
| 2 | +Shared | 2 | +IsGalaxy | 2 | +HowTo |
Who's Hiring

The Galaxy is expanding! Please help it grow.
- Responsable Opérationnel de la Plateforme de Bioinformatique ARTbio, ARTbio, Institut de Biologie Paris Seine, France
- Classification des micropolluants dans l'environnement : amélioration de l'algorithme online TyPol, INRA-LBE, Narbonne, France
- The Blankenberg Lab in the Genomic Medicine Institute at the Cleveland Clinic Lerner Research Institute is hiring postdocs.
- Galaxy Project is hiring software engineers and postdocs at Johns Hopkins, Baltimore, Maryland, United States
Have a Galaxy-related opening? Send it to outreach@galaxyproject.org and we'll put it in the Galaxy News feed and include it in next month's update.
Public Galaxy Server News
There are over 90 publicly accessible Galaxy servers and six semi-public Galaxy services. Here's what happened with them last month.
EuPathDB Galaxy Data Analysis Service
The EuPathDB Galaxy Data Analysis Service is a free Galaxy server for large-scale data analysis focused on eukaryotic pathogens.
- All EuPathDB genomes are pre-loaded.
- Pre-configured workflows are available.
- Powered by Globus Genomics
- EuPathDB tagged publications in Galaxy Publication library.
User support is available via a web form. The service is supported by EuPathDB, University of Georgia, University of Pennsylvania, and University of Liverpool.
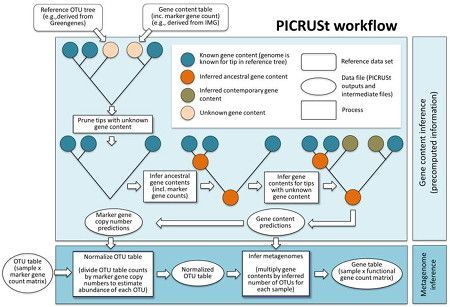
Langille Lab PICRUSt server
The Langille Lab PICRUSt server hosts PICRUSt, a bioinformatics software package designed to predict metagenome functional content from marker gene (e.g., 16S rRNA) surveys and full genomes. PICRUSt Documentation and the PICRUSt users list are available.
Anonymous access and account creation are both supported. This server is supported by the Langille Lab at Dalhousie University.
See:
- Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences, Morgan G I Langille et al. Nature Biotechnology 31, 814–821 (2013) doi:10.1038/nbt.2676
- Langille tagged publications in Galaxy Publication library.
Public Servers in Publications
We tag papers that use, mention, implement or extend public Galaxy Servers. Here are the counts for December's publications.
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 17 | >Huttenhower | 8 | >Galaxy-P | 4 | >RepeatExplorer | 4 | >Workflow4Metabolomics | |||
| 3 | >Cistrome | 1 | >Orione | 1 | >Pasteur | 1 | >EuPathDB | |||
| 1 | >APOSTL | 1 | [>OSDD MPDS](https://www.zotero.org/groups/1732893/galaxy/tags/>OSDD MPDS) | 1 | >RiboGalaxy | 1 | >RNA-Rocket | |||
| 1 | >Nebula | 1 | >PIA | 1 | >CBIB | 1 | >Langille |
Commercial Support: Intero Life Sciences and Galaxy Enterprise
We are pleased to announce a new option for commercial Galaxy support. Intero Life Sciences offers Galaxy Enterprise, a set of support services for Galaxy, including:
- Subscription/Contract Based Enterprise-Level Support
- Turn-key Customized Workflows
- Professional Services & Consulting
- Production Environment Optimization
- Data Integration Outsourcing Services
-
Enterprise Cloud Environment with SLA
- Fully managed and hosted on cloud Galaxy Enterprise with various levels of SLAs.
- Dedicated servers with dedicated secure access (VPN) including data storage and management.
- Galaxy Enterprise Certification Training
Tools
ToolShed Contributions
 ](http://toolshed.g2.bx.psu.edu/)
](http://toolshed.g2.bx.psu.edu/)Tool Shed contributions in December.
Releases
ephemeris 0.8.0
Ephemeris is a small Python library and set of scripts for managing the bootstrapping of Galaxy plugins - tools, index data, and workflows.
- Free software: Academic Free License version 3.0
- Documentation: https://ephemeris.readthedocs.org.
- Code: https://github.com/galaxyproject/ephemeris
blend4php 0.1 beta
The beta version of the blend4php package, a PHP wrapper for the Galaxy API, was released in December. It provides a PHP package for interacting with Galaxy and CloudMan. blend4php currently offers a partial implementation of the Galaxy API and includes support for datasets, data types, folder contents, folders, genomes, group roles, groups, group users, histories, history contents, jobs, libraries, library contents, requests, roles, search, tools, toolshed repositories, users, visualizations and workflows.
The motivation for development of this library is for integration with Tripal, an open-source toolkit for creation of online genomic, genetic and biological databases.
Please see the API documentation page for full information.
Earlier Releases
17.09 Galaxy Release
The Galaxy Committers published the 17.09 release of Galaxy at the end of October.
Highlights include
- Singularity Tool execution using the HPC-friendly container technology Singularity is now supported.
- Download entire collection Downloading whole collections is now possible from the history interface. (Thanks to @mvdbeek.)
- Switch tool versions in workflows You can now select exactly what version of tool you want to use when building workflows. (Thanks to @mvdbeek.)
- Security patches
- Several features were deprecated
See the 17.09 release announcement for details.
Galaxy Docker Image 17.09
The Galaxy Docker project has seen a matching release, for Galaxy 17.05. Major features include
- much improved documentation about using Galaxy Docker and an external cluster (@rhpvorderman)
- CVMFS support - mounting in 4TB of pre-build reference data (@chambm)
- Singularity support and tests (compose only)
- more work on K8s support and testing (@jmchilton)
- using .env files to configure the compose setup for SLURM, Condor, K8s, SLURM-Singularity, Condor-Docker
And
- The Galaxy Docker Project has reached more than 31k downloads on Dockerhub - not counting quay.io and all flavors
galaxy-lib 17.9.10
galaxy-lib is a subset of the Galaxy core code base designed to be used as a library. This subset has minimal dependencies and should be Python 3 compatible. It's available from GitHub and PyPi.
This revision:
- Added docs for using mulled-build with your own quay.io account (thanks to @jerowe).
- Catch errors in Conda search if nothing is found (preventing planemo-monitor from functioning properly) (thanks to @bgruening).
- Make multi-requirement container building via mulled more stable (thanks to @bgruening).
Planemo 0.47.0
Planemo is a set of command-line utilities to assist in building tools for the Galaxy project. These releases included numerous fixes and enhancements.
See GitHub for details.
Other packages that have been released in the prior 4 months.
sequence_utils 1.1.2
Galaxy's sequence utilities are a set of Python modules for reading, analyzing, and converting sequence formats. See the release notes for what's new this month.
StarForge 0.3.1-5
StarForge help build Galaxy things in virtualization:
- Build Galaxy Tool Shed dependencies
- Build Python Wheels (e.g. for the Galaxy Wheels Server)
- Rebuild Debian or Ubuntu source packages (for modifications)
These things will be built in Docker. Additionally, wheels can be built in QEMU/KVM virtualized systems. StarForge has had several updates this fall. Fixes and new features include:
- Support xz/lzma tarballs for wheel builds Pull Request 166
- Native support for auditwheel and delocate. (#160)
- Do not build sdists with the wheel subcommand by default. (#155)
- Fix a bug where the wrong working directory was set when building wheels with multiple sources. (#154)
- Fix a bug with sudo and brew install on macOS. (#151).
- Short circuit platform caching on OS X (#150).
BioBlend 0.10.0
BioBlend is a Python library for interacting with CloudMan and Galaxy‘s API. BioBlend makes it possible to script and automate the process of cloud infrastructure provisioning and scaling via CloudMan, and running of analyses via Galaxy.
See the release notes for what's new in release 0.10.0.
Pulsar 0.8.0
A Pulsar update was released in September. Pulsar is a Python server application that allows a Galaxy server to run jobs on remote systems (including Windows) without requiring a shared mounted file systems. Unlike traditional Galaxy job runners - input files, scripts, and config files may be transferred to the remote system, the job is executed, and the results are transferred back to the Galaxy server - eliminating the need for a shared file system.
This release adds several features and 4 bug fixes.
And the rest ...
Other Galaxy packages that haven't had a release in the past four months can be found on GitHub.
Other News
- Would you like to learn some Python or JavaScript and improve Galaxy at the same time? Check out our collection of very friendly issues tagged help wanted. Please feel free to ask, comment, and request guidance.
-
From Björn Grüning
shed-tools updatewill now update all Galaxy tools in your Galaxy instance. What a wonderful contribution from Ruben Vorderman!- Our community maintained CVFMS based reference data storage has now a European mirror. Thanks to JRC / EU Science Hub, the European Commission, Nate Coraor and Guy Van den Eede. This is now also available in the Galaxy Docker project! Enjoy!
-
From John Chilton
- WIP: API to build hierarchical data in Galaxy (libraries & collections) from zip files, YAML descriptions, BagIt archives, etc...
- Allow Galaxy tools to detect out of memory errors and job runners to resubmit to other resources based on this and other tool detected errors.
- Galaxy no longer depends on having samtools on its PATH. Another huge deployability step forward from Marius van den Beek!