The September 2015 Galactic News!
Welcome to the September 2015 Galactic News, a summary of what is going on in the Galaxy community. It’s been, and will be, a busy few months. If you have anything to include in the September News, please send it to [Galaxy Outreach](mailto:outreach AT galaxyproject DOT org).
New Papers
97 new papers referencing, using, extending, and implementing Galaxy were added to the Galaxy CiteULike Group in August. Highlights include:
-
FROG - Fingerprinting Genomic Variation Ontology E. Abinaya, Pankaj Narang, Anshu Bhardwaj, PLoS ONE, Vol. 10, No. 8. (5 August 2015), e0134693, doi:10.1371/journal.pone.0134693
-
EpiToolKit—a web-based workbench for vaccine design Benjamin Schubert, Hans-Philipp Brachvogel, Christopher Jürges, Oliver Kohlbacher, Bioinformatics, Vol. 31, No. 13. (01 July 2015), pp. 2211-2213, doi:10.1093/bioinformatics/btv116
-
Experimental validation of methods for differential gene expression analysis and sample pooling in RNA-seq Anto P. Rajkumar, Per Qvist, Ross Lazarus, Francesco Lescai, Jia Ju, Mette Nyegaard, Ole Mors, Anders D. Børglum, Qibin Li, Jane H. Christensen, BMC Genomics, Vol. 16, No. 1. (25 July 2015), 548, doi:10.1186/s12864-015-1767-y
-
Galaxy Integrated Omics: Web-based standards-compliant workflows for proteomics informed by transcriptomics Jun Fan, Shyamasree Saha, Gary Barker, Kate J. Heesom, Fawaz Ghali, Andrew R. Jones, David A. Matthews, Conrad Bessant, Molecular & Cellular Proteomics (12 August 2015), mcp.O115.048777, doi:10.1074/mcp.o115.048777
-
From Peer-Reviewed to Peer-Reproduced in Scholarly Publishing: The Complementary Roles of Data Models and Workflows in Bioinformatics Alejandra González-Beltrán, Peter Li, Jun Zhao, Maria S. Avila-Garcia, Marco Roos, Mark Thompson, Eelke van der Horst, Rajaram Kaliyaperumal, Ruibang Luo, Tin-Lap Lee, Tak-wah Lam, Scott C. Edmunds, Susanna-Assunta Sansone, Philippe Rocca-Serra, PLoS ONE, Vol. 10, No. 7. (8 July 2015), e0127612, doi:10.1371/journal.pone.0127612
-
Experiences with workflows for automating data-intensive bioinformatics Ola Spjuth, Erik Bongcam-Rudloff, Guillermo Carrasco C. Hernández, Lukas Forer, Mario Giovacchini, Roman Valls V. Guimera, Aleksi Kallio, Eija Korpelainen, Maciej M. Kańduła, Milko Krachunov, David P. Kreil, Ognyan Kulev, Paweł P. Łabaj, Samuel Lampa, Luca Pireddu, Sebastian Schönherr, Alexey Siretskiy, Dimitar Vassilev, Biology Direct, Vol. 10, No. 1. (19 August 2015), 43, doi:10.1186/s13062-015-0071-8
-
Challenges of Large-Scale Biomedical Workflows on the Cloud — A Case Study on the Need for Reproducibility of Results Sehrish Kanwal, Andrew Lonie, Richard O. Sinnott, Charlotte Anderson, In Computer-Based Medical Systems (CBMS), 2015 IEEE 28th International Symposium on (June 2015), pp. 220-225, doi:10.1109/cbms.2015.28
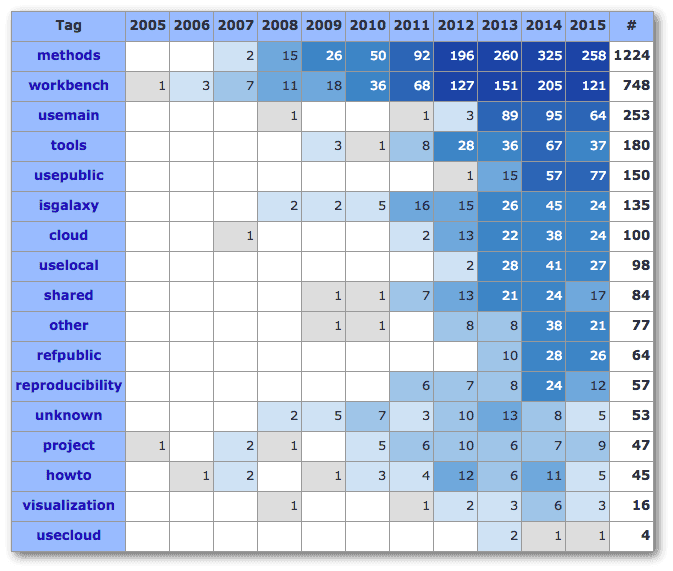
The new papers were tagged with:
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 7 | Cloud | 2 | Project | 5 | Tools | 18 | UsePublic | |||
| 1 | HowTo | 4 | RefPublic | 1 | UseCloud | 1 | Visualization | |||
| 3 | IsGalaxy | 3 | Reproducibility | 7 | UseLocal | 22 | Workbench | |||
| 57 | Methods | 2 | Shared | 12 | UseMain |
Galaxy’s First 2500 Publications
And in case you missed it, we reached 2500 publications in the Galaxy CiteULike Library. This writeup analyzes the papers by CiteULike tag and journal, across the past 11 years, and identifies some trends. Of those 2500 papers, less than 2% have a Galaxy Team member as a coauthor.
IUC Tools & Collections Hackathon
We are planning a remote hackathon on September 17th and 18th for Galaxy tool developers to hack on dataset collections.
Dataset collections enable MapReduce style workflows in Galaxy and have come a long way over the last year+. Several groups are using dataset collections and newer tools to express workflows of various degrees of complexity that were not possible in Galaxy before.
The remote nature of this gives people who don’t have the opportunity to come to GCC hackathons (which have always been productive and a lot of fun) a chance to participate in a Galaxy hackathon. Having a well defined topic will allow us to accomplish a lot and let people who don’t have particular tasks in mind find something to work on very quickly.
We are collecting ideas to work on, but we expect to attract the most participation by simply getting tool developers interested in getting help adding collection support to their existing tools and workflows to show up and participate.
If you are interested in participating in the hackathon but not interested in actual tool development - we will assemble a list of smaller, manageable Python and JavaScript tasks to work on and certainly documentation is a chronically lacking for collections so we could use help there and no actual coding would be required.
This is the first remote hackathon we have organized and we encourage ideas or advice about how to organize it please let us know. A core group of will be available on IRC all day and we will have 4 google hangouts across those days to organize, answer questions, and report progress.
Thanks,
Galaxy IUC
Galaxy Project Google Calendars
If you want to know what events are happening in the Galaxy Community, the goto place has always been the Galaxy Events page. This lists any upcoming events we know about that feature Galaxy content. This events list provides dates, locations, and contact information for each event.
There has also been the complementary Galaxy Events Google Calendar which listed events of interest to the community, whether or not they featured significant Galaxy content. To make this calendar more useful, we have split it in to two:
Galaxy Events Google Calendar ![]() This calendar now lists only events that feature Galaxy content. Pretty much, if it’s on the Events page, it’s also in this calendar.
This calendar now lists only events that feature Galaxy content. Pretty much, if it’s on the Events page, it’s also in this calendar.
Galaxy Other Events Google Calendar ![]() This lists additional events and deadlines that are relevant to the Galaxy Community, but that are not known to feature significant Galaxy content.
This lists additional events and deadlines that are relevant to the Galaxy Community, but that are not known to feature significant Galaxy content.
This split allows us to present Galaxy events in a much sparser calendar, that is more useful for setting reminders and publishing Galaxy events. And we will continue to link to past Galaxy events (and presentations, videos, examples, …) from the Events Archive.
August GalaxyAdmins Slides and Video
The next GalaxyAdmins meetup on August 20 was attended by 27 GalaxyAdmins. Aaron Petkau covered Genomic data management at Canada’s National Microbiology Laboratory with IRIDA and Galaxy and Enis Afgan presented Adding transparency and automation into the Galaxy tool installation process.
In case you missed it, slides and video are now available online.
GalaxyAdmins is a special interest group for Galaxy community members who are responsible for Galaxy installations. Our next meeting will be on October 15 where David Kovalic of Analome will present. Hope to see you there.
Upcoming Events
There are upcoming events in five countries on three continents. See the Galaxy Events Google Calendar for details on other events of interest to the community.
| |
Designates a training event offered by GTN member(s) |
Who’s Hiring
The Galaxy is expanding! Please help it grow.
- Systems Administrator IV, Fred Hutchinson Cancer Institute, Seattle, Washington, United States
- Assistant Computational Scientist at VIB, Gent, Belgium
- The Galaxy Project is hiring software engineers and post-docs
Got a Galaxy-related opening? Send it to outreach@galaxyproject.org and we’ll put it in the Galaxy News feed and include it in next month’s update.
New Public Galaxy Servers
2 new publicly accessible Galaxy servers were added in August.
Workflow4Metabolomics
- Links:
- Domain/Purpose:
- A collaborative portal dedicated to metabolomics data processing, analysis and annotation.
- Comments:
- LC-MS, GC-MS and NMR workflows are provided.
- See: Giacomoni F., Le Corguillé G., Monsoor M., Landi M., Pericard P., Pétéra M., Duperier C., Tremblay-Franco M., Martin J.-F., Jacob D., Goulitquer S., Thévenot E.A. and Caron C. (2014). Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics, doi: 10.1093/bioinformatics/btu813
- User Support:
- HowTo Tutorials
- [Help Desk](mailto:support AT workflow4metabolomics DOT org)
- Quotas:
- Account required. Anyone can request an account.
- Sponsor(s):
- See sponsor graph
EpiToolKit
- Link:
- Domain/Purpose:
- Computational immunology for the development of novel epitope-based vaccines.
- Comments:
- EpiToolKit “provides a collection of methods from computational immunology for the development of novel epitope-based vaccines including HLA ligand or potential T-Cell epitope prediction, an epitope selection framework for vaccine design, and a method to design optimal string-of-beads vaccines. Additionally, EpiToolKit provides several other tools ranging from HLA typing based on NGS data, to prediction of polymorphic peptides.”
- See EpiToolKit—a web-based workbench for vaccine design by Benjamin Schubert, Hans-Philipp Brachvogel, Christopher Jürges and Oliver Kohlbacher, Bioinformatics (2015) 31 (13): 2211-2213. doi: 10.1093/bioinformatics/btv116
- User Support:
- There is extensive documentation inlcuding step-by-step instructions.
- Quotas:
- Anyone can create an account. Alternatively, a guest account is provided with login test_user@informatik.uni-tuebingen.de and password workflowTest. Please note that the guest account shares uploaded data with all other guest users.
- Data of unregistered users and delete data will be permanently deleted after 7 days.
- Sponsor(s):
- ABI EpiToolKit is a service of the Applied Bioinformatics Group at University of Tuebingen.
Releases
Security Advisories
Two security vulnerabilities were recently discovered in Galaxy, and a third in the Galaxy Tool Shed. Because of this, the Galaxy Committers have released a number of fixes which have been rolled in to affected versions of Galaxy dating back to the 14.10 release.
- Galaxy XSS Reflection Vulnerability
- Galaxy LDAP Authentication Vulnerability
- Tool Shed Unauthorized Upload Vulnerability
Other Releases
May 2015 Galaxy Release (v 15.05) Release Notes v 15.05
BioBlend 0.6.0 and 0.6.1 BioBlend version 0.6.1 was released in July. BioBlend is a python library for interacting with CloudMan and the Galaxy API(CloudMan offers an easy way to get a personal and completely functional instance of Galaxy in the cloud in just a few minutes, without any manual configuration.) From the release CHANGELOG:
Planemo 0.13.2 Planemo is a set of command-line utilities to assist in building tools for the Galaxy project. Planemo 0.13.2 is the most recent release. See the release history.
Pulsar Pulsar 0.5.0 was released in May. Pulsar is a Python server application that allows a Galaxy server to run jobs on remote systems (including Windows) without requiring a shared mounted file systems. Unlike traditional Galaxy job runners - input files, scripts, and config files may be transferred to the remote system, the job is executed, and the results are transferred back to the Galaxy server - eliminating the need for a shared file system.
**CloudMan ** The most recent edition of CloudMan was released in August.
blend4j v0.1.2 blend4j v0.1.2 was released in December 2014. blend4j is a JVM partial reimplemenation of the Python library bioblend for interacting with Galaxy, CloudMan, and BioCloudCentral.
Galaxy Community Hubs
 |
 |
 |
| Share your training resources and experience now | Share your experience now |
One new new Galaxy Training Network member was added in August:
- University of Bradford, School of Medical Sciences, Krzysztof Poterlowicz Krzysztof offers training in Assembly, RNAseq, ChIPseq, Hi-C, DNase-Seq, and Pathway enrichment.
ToolShed Contributions
See list of tools contributed in August.