The June 2015 Galactic News!
Welcome to the June 2015 Galactic News, a summary of what is going on in the Galaxy community. These newsletters complement the Galaxy Development News Briefs which accompany new Galaxy releases and focus on Galaxy code updates.
New Papers
70 new papers referencing, using, extending, and implementing Galaxy were added to the Galaxy CiteULike Group in May. Highlights include:
- BioMAJ2Galaxy: automatic update of reference data in Galaxy using BioMAJ by Anthony Bretaudeau, Cyril Monjeaud, Yvan Le Bras, Fabrice Legeai, Olivier Collin, GigaScience, Vol. 4, No. 1. (9 May 2015), doi:10.1186/s13742-015-0063-8
- Design and bioinformatics analysis of genome-wide CLIP experiments, by Tao Wang, Guanghua Xiao, Yongjun Chu, Michael Q. Zhang, David R. Corey, Yang Xie, Nucleic Acids Research (09 May 2015), gkv439, doi:10.1093/nar/gkv439
- The Globus Galaxies platform: delivering science gateways as a service Ravi Madduri, Kyle Chard, Ryan Chard, Lukasz Lacinski, Alex Rodriguez, Dinanath Sulakhe, David Kelly, Utpal Dave and Ian Foster, Concurrency and Computation: Practice and Experience (April 2015), doi:10.1002/cpe.3486
The new papers were tagged with:
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | Cloud | - | Project | 6 | Tools | 10 | UsePublic | |||
| - | HowTo | 3 | RefPublic | - | UseCloud | 2 | Visualization | |||
| 3 | IsGalaxy | 2 | Reproducibility | 5 | UseLocal | 14 | Workbench | |||
| 38 | Methods | 3 | Shared | 10 | UseMain |
Events
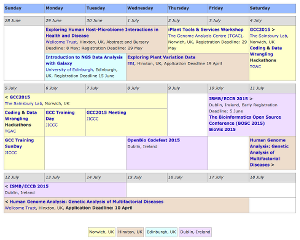
GCC2015: 4-8 July, Norwich UK
The 2015 Galaxy Community Conference (GCC2015) is the Galaxy community's annual gathering of users, developers, and administrators. Previous GCC's have drawn over 200 participants, and we expect that to happen again in 2015. GCC2015 is being hosted by The Sainsbury Lab in Norwich, UK, immediately before BOSC and ISMB/ECCB in Dublin.
There are a lot of events going on at GCC2015, including:
- Code Hackathon, 4-5 July
- Data Wrangling Hackathon, 4-5 July (new)
- Training SunDay, 5 July (new and sold out)
- Training Day, 6 July
- GCC Meeting, 7-8 July
Registration is Open!
registration is open, and there is still available space for most GCC2015 events. If you work in data-intensive life science research, then there is no meeting that will be more relevant this year than GCC2015. We look forward to seeing you there.
Late Poster Abstracts
There are still a limited number of poster slots available and we welcome poster abstract submissions. Poster authors will be notified of acceptance status within two weeks. Please consider presenting your work. If you are dealing with big biological data, then this meeting wants to hear about it.
You can also submit late oral presentation submissions. These will be considered as cancellations happen (and they usually do).
Other Events Near GCC2015
Looking to extend your trip? There are also at least 7 other events going on in the British Isles in the week before and week after GCC2015, including events
In Norwich
In Cambridge
- Exploring Human Host-Microbiome Interactions in Health and Disease
- Exploring Plant Variation Data
- Human Genome Analysis: Genetic Analysis of Multifactorial Diseases
In Dublin
- OpenBio Codefest 2015
- ISMB / EECB 2015 including BOSC 2015 and BioVis 2015 and lots of Galaxy content.
See the Events Aplenty Near GCC2015 news item for dates and deadlines.
GCC2015 Sponsorships
Hackathons Sponsor: Johns Hopkins University Data Science Specialization Program
We are pleased to announce that the Johns Hopkins University Data Science Specialization Program offered on Coursera is the exclusive sponsor for both the GCC2015 Coding Hackathon and the Data Wrangling Hackathon.
This program includes a Genomic Data Science course covering
- Basic computational, biological, and statistical skills for analyzing big genomic data
- Foundational tools for successfully engaging in this rapidly changing field
- How to go from unprocessed next generation sequencing data to meaningful biological results
and introduces Galaxy, Bioconductor, Python, command line, and many other tools.
The series of nine MOOCs are now open for enrollment and free to anyone.
Training Day Sponsor: Amazon Web Services
We are pleased to again have Amazon Web Services (AWS) as the sponsor for the GCC2015 Training Days. This is the 4th year in a row AWS has sponsored GCC Training Days. Many of the sessions will be using AWS directly, or servers running on AWS.
Call for Sponsors
The 2015 Galaxy Community Conference (GCC2015) is still accepting Sponsorships. Your organisation can play a prominent part in the Galaxy community by sponsoring GCC2015. Sponsorship is an excellent way to raise your organization’s visibility.
Several sponsorship levels are available, including two levels of premier sponsorships that include presentations. Premium sponsorships are limited, however, so you are encouraged to act soon.
Please let the organisers know if you are interested in helping make this event a success.
June GalaxyAdmins Meetup
Please join us for the next GalaxyAdmins meetup on June 18 when Peter Briggs of the University of Manchester will speak on
Galactic Engineering: Experiences deploying Galaxy and developing tools within the Bioinformatics Core Facility at the University of Manchester
GalaxyAdmins will also have an in-person meetup at GCC2015.
GalaxyAdmins is a special interest group for Galaxy community members who are responsible for Galaxy installations.
Other Events
There are upcoming events in seven countries on three continents. See the Galaxy Events Google Calendar for details on other events of interest to the community.
| |
Designates a training event offered by GTN member(s) |
Who's Hiring
The Galaxy is expanding! Please help it grow.
- Post-Doc CDD-IR CEA-Institut de Génomique, Evry, France Application deadline extended to 2015/06/30
- Computational Metabolomics Professor, Penn State University, Pennsylvania, United States
- The Galaxy Project is hiring software engineers and post-docs
Got a Galaxy-related opening? Send it to outreach@galaxyproject.org and we'll put it in the Galaxy News feed and include it in next month's update.
New Public Galaxy Servers
The VirAmp server was added to the list of public Galaxy servers in May:
VirAmp
VirAmp is a web-based semi-de novo fast virus genome assembly pipeline designed for extremely high coverage NGS data. From VirAmp: a galaxy-based viral genome assembly pipeline, Yinan Wan, Daniel W Renner, Istvan Albert and Moriah L Szpara, GigaScience 2015, 4:19 doi:10.1186/s13742-015-0060-y VirAmp "combines existing tools and techniques and presents them to end users via a web-enabled Galaxy interface. Our pipeline allows users to assemble, analyze, and interpret high coverage viral sequencing data with an ease and efficiency that was not possible previously. Our software makes a large number of genome assembly and related tools available to life scientists and automates the currently recommended best practices into a single, easy to use interface. We tested our pipeline with three different datasets from human herpes simplex virus (HSV)."
VirAmp includes Documentation and is supported by the Huck Institutes of the Life Sciences and the Department of Biochemistry and Molecular Biology, and Pennsylvania State University
Releases
May 2015 Galaxy Release (v 15.05)
The May 2015 Galaxy Release is out! Release Notes v 15.05
Release Highlights
- Authentication Plugins - Galaxy now has native support for LDAP and Active Directory via a new community developed authentication plugin system.
- Tool Sections - Tool parameters may now be groupped into collapsable sections.
- Collection Creators - New widgets have been added that allow much more flexibility when creating simple dataset pair and list collections.
- And much more
The release notes have a new home at Read the Docs. We hope that you like the new format as much as we do!
Thanks for using Galaxy!
The Galaxy Team
Pulsar 0.5.0
Pulsar 0.5.0 was released in May. Enhancements include:
- Allow cURL downloader to resume transfers during staging in (thanks to Nate Coraor). 0c61bd9
- Fix to cURL downloaders status code handling (thanks to Nate Coraor). 86f95ce
- Fix non-wheel installs from PyPI. Issue 72
- Fix mesos imports for newer versions of mesos (thanks to Kyle Ellrott). fe3e919
- More, better logging. 2b3942d, fa2b6dc
Pulsar is a Python server application that allows a Galaxy server to run jobs on remote systems (including Windows) without requiring a shared mounted file systems. Unlike traditional Galaxy job runners - input files, scripts, and config files may be transferred to the remote system, the job is executed, and the results are transferred back to the Galaxy server - eliminating the need for a shared file system.
Planemo 0.12.2
Planemo saw four major and several minor releases this past month. The most recent is v0.12.2. See the release history for full details and links.
Planemo is a set of command-line utilities to assist in building tools for the Galaxy project.
Others
**CloudMan ** The most recent edition of CloudMan was released in August.
BioBlend V0.5.3
BioBlend v0.3 was also released in March.
BioBlend is a python library for interacting with CloudMan and the Galaxy API. (CloudMan offers an easy way to get a personal and completely functional instance of Galaxy in the cloud in just a few minutes, without any manual configuration.)
blend4j v0.1.2 blend4j v0.1.2 was released in December 2014. blend4j is a JVM partial reimplemenation of the Python library bioblend for interacting with Galaxy, CloudMan, and BioCloudCentral.
Other News
-
From John Chilton:
- EDAM (edamontology.org) format added to all Galaxy datatypes. Huge thanks to Pasteur team & Helena Rasche!
- Connecting a dockerized Galaxy container to a Condor cluster pretty awesome stuff from Devon Ryan.
- Just merged collaborative work spearheaded by Helena Rasche adding RStudio integration to Galaxy
- Videos from Pitagora Galaxy workshop are now online
- GigaScience Pushes Metabolomics Open Data & Training, by Scott Edmunds