February 2019 Galactic News
Events (GCC Training, GalaxyAdmins), Platforms, Pubs, Jobs, COTM!, Doc, Training, Tools, Releases and more
The February 2019 Galactic News is here! This is a summary of what is going on in the Galaxy community.
-
- Voting on GCC2019 training topics open through February 4th!.
- ELIXIR Galaxy Community Workshop pre-registration is open!
- GalaxyAdmins needs your topic suggestions.
- 160 new publications, great resources lead to great insight.
- Some most excellent Galaxy Platform News, including proteo-transcriptomics, RNA analyis tools, Genome Browsers, and more!
- The Tutorial of the Month is ... Genome annotation with Prokka!
- At least 23 Open positions in 6 countries on 2 continents.
- Doc, Hub, and Training Updates, including GO enrichment analysis and Age prediction using machine learning.
- ToolShed contributions.
- New releases of galaxy-lib and Planemo.
- And a bunch of other news too.
If you have anything to add to next month's newsletter, then please send it to outreach@galaxyproject.org.
Event News
GCC2019 Training Topic Voting Is Open
The 2019 Galaxy Community Conference (GCC2019) will be held in Freiburg, Germany, 1-6 July. Like previous Galaxy Community Conferences, GCC2019 will feature invited keynotes, accepted talks, posters, demos, birds-of-a-feather gatherings, and training. The format of GCC2019 will be a bit different than the previous years: it starts with 1 day of training and then 3 days with more specialized training sessions aligned with talks. You can read more about the new format on the conference FAQ.
The GCC2019 Organizing Committee compiled training topics nominated by the community and expanded this list with community requests and topics from the previous GCC to offer a broad range of training sessions to be voted on by the community for GCC2019. Voting closes February 4th.
Training topics that are offered at GCC2019 are determined by the community so BY YOU!
You can vote for as many topics as you want, but please note that the more topics you vote for the less your vote for each one counts. What? Here's an example. If Moni votes for 4 topics then each of her votes counts for 1/4 of a point. If Dave votes for 22 topics then each of his votes counts for 1/22 of a point.
Please take a moment and vote on which training topics you would like to see at GCC2019, your vote matters!
ELIXIR Galaxy Community Workshop
Join the Galaxy Community of Elixir for a three day workshop at Station Biological de Roscoff, France to participate in workshops and hackathons focused on tool implementation in Galaxy, development of training materials for the Galaxy Training Network (GTN), and how to teach using GTM materials.
During the first 1 1/2 days, the attendees will learn about the integration of high-quality tools within Galaxy with their dependencies (Bioconda, Planemo) using the IUC best practice guidelines. They will also learn how to use Galaxy as a training tool and develop training material for the Galaxy Training Network. The second half of the workshop will be dedicated to Hackathon sessions where attendees will be able to bring their own projects around tool integration and/or training material and develop them collaboratively, with the support of community experts. Additional information can be found here.
Find pre-registration here.
GalaxyAdmins needs your topic suggestions
After a two year hiatus, the bimonthly online GalaxyAdmins meetups will return on February 28. GalaxyAdmins is a discussion group for Galaxy community members who are responsible for Galaxy installations. Our online meetups are around an hour long and feature a presentation followed by an open discussion. It's a great place to catch up on what your fellow admins are thinking about.
But, before we start meeting this month, we need to know what you care about. What do you want to learn? Who do you want to hear from? What's causing you trouble? Or, what have you worked that your fellow GalaxyAdmins would benefit from knowing?
Got some ideas? Nominate them
Upcoming events
These and other Galaxy related events are coming up in the next few months:
Publications
160 new publications referencing, using, extending, and implementing Galaxy were added to the Galaxy Publication Library in January.
Highlighted Publications
Galactic and Stellar publications from January:
- G-OnRamp: A Galaxy-based platform for creating genome browsers for collaborative genome annotation, Yating Liu, Luke Sargent, Wilson Leung, Sarah C.R. Elgin, Jeremy Goecks. bioRxiv 499558; doi: https://doi.org/10.1101/499558
- Galaxy mothur Toolset (GmT): a user-friendly application for 16S rRNA gene sequencing analysis using mothur, Saskia D Hiltemann, Stefan A Boers, Peter J van der Spek, Ruud Jansen, John P Hays, Andrew P Stubbs. GigaScience, giy166, doi:10.1093/gigascience/giy166
- MEGARes: an antimicrobial resistance database for high throughput sequencing, Steven M. Lakin, Chris Dean, Noelle R. Noyes, Adam Dettenwanger, Anne Spencer Ross, Enrique Doster, Pablo Rovira, Zaid Abdo, Kenneth L. Jones, Jaime Ruiz, Keith E. Belk, Paul S. Morley, Christina Boucher. Nucleic Acids Research, Volume 45, Issue D1, 4 January 2017, Pages D574–D580, doi: 10.1093/nar/gkw1009
- Peptimapper: proteogenomics workflow for the expert annotation of eukaryotic genomes, Laetitia Guillot, Ludovic Delage, Alain Viari, Yves Vandenbrouck, Emmanuelle Com, Andrés Ritter, Régis Lavigne, Dominique Marie, Pierre Peterlongo, Philippe Potin and Charles Pineau. BMC Genomics 201920:56 doi:10.1186/s12864-019-5431-9
- Growing and cultivating the forest genomics database, TreeGenes, Taylor Falk, Nic Herndon, Emily Grau, Sean Buehler, Peter Richter, Sumaira Zaman, Eliza M Baker, Risharde Ramnath, Stephen Ficklin, Margaret Staton, Frank A Feltus, Sook Jung, Doreen Main, Jill L Wegrzyn. Database, Volume 2018, 1 January 2018, bay084, doi: 10.1093/database/bay084
- Exploring the Effects of Spaceflight on Mouse Physiology using the Open Access NASA GeneLab Platform, Afshin Beheshti, Yasaman Shirazi-Fard, Sungshin Choi, Daniel Berrios, Samrawit G. Gebre, Jonathan M. Galazka, Sylvain V. Costes. Journal of Visual Experiments (143), e58447, doi:10.3791/58447 (2019)
- Mind the gaps: overlooking inaccessible regions confounds statistical testing in genome analysis, Diana Domanska, Chakravarthi Kanduri, Boris Simovski and Geir Kjetil Sandve. BMC Bioinformatics 201819:481, doi: 10.1186/s12859-018-2438-1
- VAPPER: High-throughput Variant Antigen Profiling in African trypanosomes, Sara Silva Pereira, John Heap, Andrew R Jones, Andrew P. Jackson. bioRxiv 492074; doi: 10.1101/492074
- PrediTALE: A novel model learned from quantitative data allows for new perspectives on TALE targeting, Annett Erkes, Stefanie Mücke, Maik Reschke, Jens Boch, Jan Grau. bioRxiv 522458; doi: 10.1101/522458
- Enhancing Access to Digital Media: The Language Application Grid in the HTRC Data Capsule, James Pustejovsky, Marc Verhagen, Keongmin Rim, Yu Ma, Liang Ran, Samitha Liyanage, Jaimie Murdock, Robert H. McDonald, Beth Plale. In PEARC17 Proceedings of the Practice and Experience in Advanced Research Computing 2017 on Sustainability, Success and Impact, Article No. 60. doi: 10.1145/3093338.3104171
- Predicting runtimes of bioinformatics tools based on historical data: Five years of Galaxy usage, Anastasia Tyryshkina, Nate Coraor, Anton Nekrutenko. Bioinformatics, btz054, doi: 10.1093/bioinformatics/btz054
- Tools for Understanding miRNA–mRNA Interactions for Reproducible RNA Analysis, Andrea Bagnacani, Markus Wolfien, Olaf Wolkenhauer. In: Lai X., Gupta S., Vera J. (eds) Computational Biology of Non-Coding RNA. Methods in Molecular Biology, vol 1912. Humana Press, New York, NY
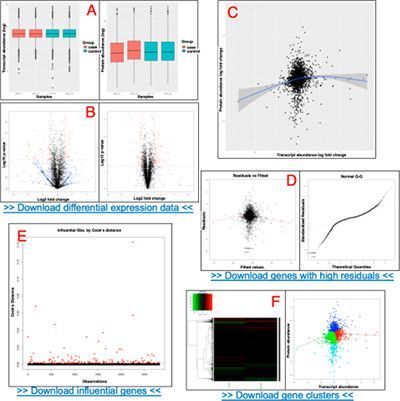
- QuanTP: A Software Resource for Quantitative Proteo-Transcriptomic Comparative Data Analysis and Informatics, Praveen Kumar, Priyabrata Panigrahi, James Johnson, Wanda J. Weber, Subina Mehta, Ray Sajulga, Caleb Easterly, Brian A. Crooker, Mohammad Heydarian, Krishanpal Anamika, Timothy J. Griffin , and Pratik D. Jagtap. Journal of Proteome Research, Article ASAP DOI: 10.1021/acs.jproteome.8b00727
- miCloud: A Plug-n-Play, Extensible, On-Premises Bioinformatics Cloud for Seamless Execution of Complex Next-Generation Sequencing Data Analysis Pipelines, Baekdoo Kim, Thahmina Ali, Changsu Dong, Carlos Lijeron, Raja Mazumder, Claudia Wultsch, and Konstantinos Krampis. Journal of Computational Biology doi: 10.1089/cmb.2018.0218
Ten of the fourteen highlighted publications are open access.
Publication Topics
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 111 | +Methods | 47 | +UsePublic | 23 | +UseMain | 16 | +Workbench | |||
| 11 | +UseLocal | 10 | +RefPublic | 10 | +IsGalaxy | 9 | +Tools | |||
| 6 | +Reproducibility | 3 | +Other | 2 | +Cloud | 2 | +HowTo | |||
| 2 | +Shared | 2 | +Project |
Galaxy Platforms News
The Galaxy Platform Directory lists resources for easily running your analysis on Galaxy, including publicly available servers, cloud services, and containers and VMs that run Galaxy. Here's what's been added since last month:
QuanTP
QuanTP is a Docker image containing a fully-operational Galaxy instance with pre-installed QuanTP, a suite of tools and functionalities critical for proteo-transcriptomics, including statistical algorithms for assessing the correlation between single transcript–protein pairs as well as across two cohorts, outlier identification and clustering, along with a diverse set of results visualizations.
QuanTP is available in this Docker image, and on the Proteogenomics Gateway and UseGalaxy.eu. It has an Installation guide, and Supporting information, including Instructions for accessing the QuanTP tool on Jetstream, Instructions for accessing and running the Mouse data, and Accessing the QuanTP on the usegalaxy.eu. QuanTP is supported by the University of Minnesota, Minneapolis.
miCloud
miCloud is a Docker Image that leverages cloud technologies to provide an easy-to-use streamlined NGS data analysis using local computational hardware. It includes three preconfigured and ready-to-execute NGS pipelines: two for single or paired-end ChIP-Seq data and one for paired-end RNA-Seq data. miCloud can also provide access to a large selection of ready-to-execute bioinformatics pipelines, available from online Docker repositories such as BioContainers, DockStore, or BioShadock. It provides a workflow-centric interface to a Galaxy engine, using BioBlend.
miCloud is supported by Weill Cornell Medicine and Department of Biological Sciences, Hunter College of the City University of New York.
RNA Workbench
RNA Workbench, in addition to being available as a Docker container, is also available as public server hosted at UseGalaxy.eu. RNA Workbench has a collection of over 50 tools dedicated to different research areas of RNA biology, including RNA structure analysis, RNA alignment, RNA annotation, RNA-protein interaction, ribosome profiling, RNA-Seq analysis, and RNA target prediction. This server has been up for a while, but we are just adding it to the platforms directory.
The workbench is developed by the RNA Bioinformatics Center (RBC). This center is one of the eight service units of the German Network for Bioinformatics Infrastructure, running the German ELIXIR Node.
Hi-C Explorer
Hi-C Explorer is another server that has been up for a while, but is also new to the Galaxy Platform Directory. It facilitates the study of the 3D conformation of chromatin by allowing Hi-C data processing, analysis and visualization. See the "Hi-C analysis of Drosophila melanogaster cells using HiCExplorer tutorial and example history.
Hi-C Explorer is also hosted at UseGalaxy.eu.
G-OnRamp
G-OnRamp provides a user-friendly, web-based platform for collaborative, end-to-end annotation of eukaryotic genomes using UCSC Assembly Hubs and JBrowse/Apollo genome browsers with evidence tracks derived from sequence alignments, ab initio gene predictors, RNA-Seq data, and repeat finders. It is available as a virtual machine that can be deployed locally, and as an Amazon Machine Image that can be run on AWS and launched with CloudLaunch.
Use the G-OnRamp output to visualize large genomics datasets, and can utilize the output to drive collaborative genome annotation projects in both research and educational settings. It has documentation:
- How to use G-OnRamp
- Learn G-OnRamp
- Virtual Machine Installation Walkthrough
- CloudLaunch Deployment Walkthrough (using the AMI)
G-OnRamp is supported by Washington University in St. Louis and Oregon Health & Science University
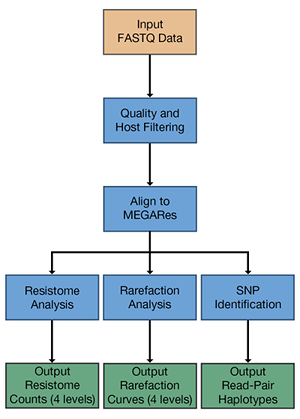
AmrPlusPlus
AmrPlusPlus is a Docker container with a Galaxy-based metagenomics pipeline that is intuitive and easy to use. AmrPlusPlus takes advantage of current and new tools to help identify and characterize resistance genes within metagenomic sequence data. The pipeline can be used under a local instance of Galaxy and installed via Galaxy's Main Tool Shed. It is also available as a Galaxy-based Docker Image, using base images developed by Björn Grüning at the University of Freiburg. It has Documentation and Email support. AmrPlusPlus is supported by Colorado State University.
Lifeportal
Lifeportal is a Galaxy server available to any researcher in Norway (and possibly any other user willing to use the service). LifePortal implements about 400 applications. It runs Galaxy version 18.09 and jobs are executed on the Abel cluster (~650 nodes) using the slurm-drmaa library. User support is available via email.
Every user with their own project in Lifeportal can manage it through the menus built in the Galaxy GUI. Lifeportal is supported by University of Oslo, Norwegian Bioinformatics Platform, MLS UiO, and UninetT Sigma2
LAPPS Grid
THIS CONTENT HAS BEEN UPDATED SINCE ORIGINAL PUBLICATION.
And from this cool writeup (that has nothing to do with life science): Learning from data: Using technology to gain deeper meaning from archives, by Emily Mercer of San Jose State University, describing work by James Pustejovsky of Brandeis University:
One example is the collaboration between the American Archive of Public Broadcasting and James Pustejovsky, professor at Brandeis University. In this collaboration James and his research team have created a software package using the Galaxy Workflow Engine and Language Application Grid ([LAPPS Grid](/use/lapps-grid/)) for automatic extraction of metadata from video assets using machine learning methodologies (Oomen et al., 2018; [Pustejovsky et al., 2017](https://doi.org/10.1145/3093338.3104171)). What was once a time exhaustive process of real-time viewing and transcribing of film and video is now as simple as uploading a video file and applying a workflow. Processing takes seconds and afterwards the user has access to the metadata, for example, the text captions on the screen. This metadata can be exported as PBCore, a metadata standard specific to public broadcasting (Oomen et al., 2018; Pustejovsky et al., 2017).
There's also some Jetstream in the mix too.
Galaxy Platforms in Publications
We tag papers that use, mention, implement or extend public Galaxy platforms (servers, services, clouds, containers...). Here are the counts for the past month's publications:
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 23 | >Huttenhower | 6 | >RepeatExplorer | 5 | >ARGs-OAP | 3 | >ABiMS | |||
| 3 | >Cistrome | 3 | >Galaxy-P | 3 | >UseGalaxy.eu | 2 | [>LAPPS Grid](https://www.zotero.org/groups/1732893/galaxy/tags/>LAPPS Grid) | |||
| 2 | [>RNA Workbench](https://www.zotero.org/groups/1732893/galaxy/tags/>RNA Workbench) | 2 | >UseGalaxy.org | 1 | >AmrPlusPlus | 1 | >ASH | |||
| 1 | >G-OnRamp | 1 | >GeneLab | 1 | [>Genomic Hyperbrowser](https://www.zotero.org/groups/1732893/galaxy/tags/>Genomic Hyperbrowser) | 1 | >GIO | |||
| 1 | >GmT | 1 | >GVL-MEL | 1 | >GVL-QLD | 1 | [>Halogen Bonding](https://www.zotero.org/groups/1732893/galaxy/tags/>Halogen Bonding) | |||
| 1 | >HiCExplorer | 1 | [>Martin Luther](https://www.zotero.org/groups/1732893/galaxy/tags/>Martin Luther) | 1 | >miCloud | 1 | >Peptimapper | |||
| 1 | >PRABI | 1 | >QuanTP | 1 | >SouthGreen | 1 | >TDPortal | |||
| 1 | >TreeGenes | 1 | >Trinity | 1 | >UseGalaxy.org | 1 | >Workflow4Metabolomics |
New Galactic Blog Posts
 ](/blog/2019-01-totm/)
](/blog/2019-01-totm/)
There are two new Galactic Blog entries:
-
Tutorial of the Month: Genome annotation with Prokka, selected by Simon Gladman:
- read about using Prokka for annotation of draft genome sequences
- by Bérénice Batut
-
Hey Galaxy People! Want easy proteo-transcriptomics!??!
- an enthusiastic post by Ben Orsburn about QuanTP
Who's Hiring

The dark energy of irreproducible research is threatening the science universe! Please help the Galaxy push it back!
- Genomic data analyst, ARTbio, Institut de Biology Paris Seine, campus Jussieu of the Sorbonne-Université, Paris, France
- Cloud Engineer V, Fred Hutchinson Cancer Research Center, Seattle, Washington, United States.
-
ELIXIR Belgium has four Galaxy-related openings in Ghent:
- ELIXIR Open Science Community Manager, VIB-UGent Center for Plant Systems Biology
- ELIXIR Software developer data management tools, VIB-UGent Center for Plant Systems Biology
- ELIXIR Bioinformatics Trainer, VIB Bioinformatics Core
- ELIXIR Scientific Cloud Coordinator, VIB-UGent Center for Plant Systems Biology
- Software Developer, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, United States. "Basic automated analysis workflows using Galaxy for 16S marker gene, metagenomic, and metatranscriptomic data leveraging existing software."
- M2 - bio-informatique, Laboratoire de Virologie , Hôpital St Louis, Paris, France.
- Bioinformatics Scientist, Providence Health Services, Portland, Oregon, United States. Looking for someone "fluent in Galaxy or willing to learn"
- Etude et galaxyfication d’un pipeline de traitement de données en lipidomique, Centre INRA Auvergne-Rhône-Alpes, Theix, France
- Cloud Computing Bioinformatics Programmer working with IRIDA, Simon Fraser University, Vancouver, Canada
- Senior IT DevOps Data Engineer and Senior IT Data Engineer (IT Bioinformatician), Adaptimmune, Abingdon, Oxfordshire, United Kingdom. Required: Experience with following bioinformatic pipeline tools: Galaxy…
- Specialist Solutions Architect -- Biological Sciences, Amazon Web Services, United States
- Principal Technical Business Development Manager: AWS Research in Biomedical, Amazon Web Services, United States
- Bioinformatics Scientist, ResearchDx, Irvine, California, United States
- NGS Scientific Applications Specialist, Integrated DNA technologies, Iowa City, Iowa, United States
-
The The European Galaxy Team has open positions, Freiburg, Germany
- Software engineer, system analysts/administrators, data analyst, and a comunnity and/or research manager
- The Blankenberg Lab in the Genomic Medicine Institute at the Cleveland Clinic Lerner Research Institute is hiring postdocs.
- Galaxy Project is hiring two software engineers at Johns Hopkins University, Baltimore, Maryland, United States.
Have a Galaxy-related opening? Send it to outreach@galaxyproject.org and we'll put it in the Galaxy News feed and include it in next month's update.
Doc, Hub, and Training Updates
Updates from the Galaxy Training Materials in January, and the Hub as well:
GTN Training Materials
- New Age prediction using machine learning, by Anup Kumar.
- New GO enrichment analysis, by igcbioinformatics, Maria Doyle, and dsobral.
- Join the February 2019 GTN CoFest. Held on 21 February, the CoFest will be coordinated via Hangout (drop-in channel will be open the whole day) and the GTN Gitter channel.
Hub
ToolShed Contributions
 ](http://toolshed.g2.bx.psu.edu/)
](http://toolshed.g2.bx.psu.edu/)Tool Shed contributions in January 2019.
Releases
New additions to the Galaxy Ecosystem.
galaxy-lib 18.9.2
galaxy-lib is a subset of the Galaxy core code base designed to be used as a library. This subset has minimal dependencies and should be Python 3 compatible. It's available from GitHub and PyPi.
Planemo 0.58.1
Planemo is a set of command-line utilities to assist in building tools for the Galaxy project. These releases included numerous fixes and enhancements.
See GitHub for details.
Other News
- The Galaxy statistics page has received it's semi-annual update.
-
From James Taylor:
- Visualizing scRNA-Seq data using cellxgene inside Galaxy.
PAG 2019 Presentations and Posters are Available
Copies of most of the Galaxy related presentations and posters form the Plant and Animal Genome Conference are now online. Many thanks to the all the presenters for sharing their during and after the conference.