November 2014 Galaxy Update
Welcome to the November 2014 Galaxy Update, a summary of what is going on in the Galaxy community. Galaxy Updates complement the Galaxy Development News Briefs which accompany new Galaxy releases and focus on Galaxy code updates.
Thanks!
Thanks to everyone who took the Galaxy Questionnaires in October. We received 155 responses, which is an amazing number. We will use this input to help prepare the next grant for the Galaxy Project. Thanks again for your time and thoughtful responses.
IRC Channel is Now Publicly Archived
The #galaxyproject IRC channel now has an online public archive. These archives have also been included in the Galaxy search engine. The archive started on October 22, 2014.
This was proposed and then discussed, and approved on Galaxy Biostar.
Galaxy Training Network
The Galaxy Training Network (GTN) was launched October 16 with 16 charter member organizations. The GTN is a network of trainers who teach bioinformatics using Galaxy, or teach about Galaxy itself. The GTN aims to make it easy to find Galaxy trainers, and to share and discover the wealth of training resources available for Galaxy. This includes training materials, a trainer directory, best practices, and guidance on computing platforms for teaching with Galaxy. The Galaxy Training Network is accessible to the entire community.
If you teach with Galaxy, then please consider adding your organization, materials, and best practices. Since the GTN launched two weeks ago, 4 new organizations have joined:
- Alberta Children's Hospital Research Institute, Calgary, Canada
- Memorial Sloan Kettering Cancer Center, Rätsch Laboratory, New York City, United States
- MMG@IICT, Hyderabad, India
- South Green Platform, Montpellier, France
This brings the total to 20 training organizations on 5 continents.
Events
Galaxy Days: 2-3 December, Paris
The French Working Group GALAXY-IFB (Institut Français de Bioinformatique) is organizing a second session around the Galaxy portal. The event will be at Institut Curie in Paris over two days. This year, we want to involve two communities: biologists (also known as Galaxy 'users') and bioinformaticians (Galaxy 'developers'). The goal is to present user experience around the portal, from a single user to a wider community:
- Dec 2 (13:30-17:30): Galaxy's user experiences, and discussion on how the platform is (or is not) useful for building analysis.
- Dec 3 (09:00-17:00): Technology talks (new environment, Galaxy in production, ...)
Interested? Please contact [ifb DOT galaxy AT sb DASH roscoff DOT fr](mailto:ifb DOT galaxy AT sb DASH roscoff DOT fr) for more information.
The French IFB Galaxy Working Group:
URGI, GenoToul, MIGALE, PFEM, SouthGreen, Institut Curie, ABiMS
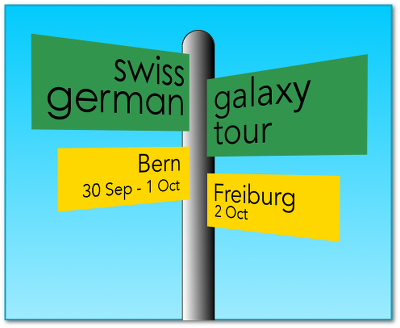
Swiss German Galaxy Tour 2014 Report
After the big success of the first Swiss Galaxy Workshop two years ago, we decided to organize a similar event again this fall. This time, we added a training day prior to the workshop, and a developer day after the workshop. The first two days were held in Bern (Switzerland), and the third in Freiburg (Germany). Hence we called the whole event: "Swiss German Galaxy Tour 2014".
More than 40 people registered for the event, signing up for one, two or all three days, ...
- read more -
Fall 2014 GUGGO Events Report
Three events were sponsored by the Galaxy User Group Grand Ouest (GUGGO) in western France earlier this fall. Summaries of all 3 events are now available online.
The Tools integration on Galaxy Workshop was held 11 September. The summary includes ...
- read more -
Other Events
There are upcoming events in France, Germany, Australia, Italy, and the United States. See the Galaxy Events Google Calendar for details on other events of interest to the community.
| Date | Topic/Event | Venue/Location | Contact |
|---|---|---|---|
| November 3-5 | Des bonnes pratiques d'intégration d'outils sous Galaxy Workshop full, but you can get on the waiting list |
Station Biologique de Roscoff, France | Christophe Caron |
| November 3-5 | Galaxy NGS Training in the Group of Prof. Dr. Bettina Kempkes Workshop full, but you can get on the waiting list |
Helmholtz Zentrum München, Germany | Björn Grüning |
| November 3-6 | Galaxy training days Workshop full, but you can get on the waiting list |
INRA de Toulouse Midi-Pyrénées, France | GenoToul Bioinformatics Team |
| November 5-8 | Rapidly bringing software to biologists with Galaxy and Docker | Biological Data Science, Cold Spring Harbor Laboratory, New York, United States | John Chilton |
| Building Galaxy Japan community (See Pitagora Galaxy) |
Ryota Yamanaka | ||
| November 16 | Deciphering Big Data Stacks: An Overview of Big Data Tools | Workshop on Big Data Analytics: Challenges, and Opportunities (BDAC-14), Supercomputing 2014 (SC14), New Orleans, Louisiana, United States | Enis Afgan |
| November 18-20 | Analisi dati Next Generation Sequencing con Galaxy | Cagliari, Italy | CRS4 |
| November 19-20 | Workshop: Extended RNA-Seq analysis | The University of Queensland, Brisbane, Queensland, Australia | Mark Crowe |
| November 21 | Galaxy Cluster to Cloud - Genomics at Scale | GCE: The 9th Gateway Computing Environments Workshop, Supercomputing 2014 (SC14), New Orleans, Louisiana, United States | Enis Afgan |
| November 26-28 | RNA-Seq & ChIP-Seq analysis course using Galaxy | PRABI, Lyon, France | Navratil V., Oger C., Veber P., Deschamps C., Perriere G. |
| December 2-3 | Galaxy Day | Institut Curie, Paris, France | IFB Galaxy |
| December 5-8 | Next Generation Data Analysis Workshop | UC Riverside, Riverside, California, United States | Rakesh Kaundal |
| December 9-11 | Microarray data analysis on Galaxy | BIRD IFB core facility Nantes University/INSERM, Nantes, France | Raluca Teusan, Audrey Bihouée, Edouard Hirchaud |
| December 16-19 | RNA-Seq and ChIP-Seq Analysis with Galaxy | UC Davis, California, United States | UC Davis Bioinformatics Training |
| 2015 | |||
| January 10-14 | Galaxy for SNP and Variant Data Analysis | Plant and Animal Genome XXIII (PAG2014), San Diego, California, United States | Dave Clements |
| February 9-13 | Analyse bioinformatique de séquences sous Galaxy | Montpellier, France | J.F. Dufayard |
| February 16-18 | Managing and Disseminating Tools and Data in Galaxy | Genome and Transcriptome Analysis, part of Molecular Medicine Tri-Conference, San Francisco, California, United States | James Taylor |
| July 6-8 | 2015 Galaxy Community Conference (GCC2015) | The Sainsbury Lab, Norwich, United Kingdom | Galaxy Outreach |
New Papers
38 papers were added to the Galaxy CiteULike Group in October, including:
- Flexible and accessible workflows for improved proteogenomic analysis using the Galaxy framework, by Pratik D. Jagtap, James E. Johnson, Getiria Onsongo, et al., J. Proteome Res. (10 October 2014), doi:10.1021/pr500812t
- iReport: a generalised Galaxy solution for integrated experimental reporting, by Saskia Hiltemann, Youri Hoogstrate, Peter van der Spek, Guido Jenster, Andrew Stubbs, GigaScience, Vol. 3, No. 1. (2014), 19, doi:10.1186/2047-217x-3-19
- ExomeAI: Detection of recurrent Allelic Imbalance in tumors using whole Exome sequencing, by Javad Nadaf, Jacek Majewski, Somayyeh Fahiminiya, Bioinformatics (08 October 2014), btu665, doi:10.1093/bioinformatics/btu665
The new papers were tagged in these areas:
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 4 | Cloud | - | Project | 4 | Tools | 3 | UsePublic | |||
| - | HowTo | 3 | RefPublic | - | UseCloud | - | Visualization | |||
| 3 | IsGalaxy | 1 | Reproducibility | 2 | UseLocal | 8 | Workbench | |||
| 19 | Methods | 4 | Shared | 6 | UseMain |
Who's Hiring
The Galaxy is expanding! Please help it grow.
- Bioinformatician, Max Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany
- Research Specialist, Michigan State University, United States
- Galaxy Workflow Developer, John Innes Centre, Norwich, United Kingdom. Closes Nov 5.
- Computational Science Developer I, Cold Spring Harbor Laboratory (CSHL), New York, United States
- Statistical Genomics Postdoc opening in the Makova lab at Penn State
- The Galaxy Project is hiring software engineers and post-docs
Got a Galaxy-related opening? Send it to outreach@galaxyproject.org and we'll put it in the Galaxy News feed and include it in next month's update.
New Public Servers
Two new public Galaxy servers were added to the published list in October:
Majewski Lab Galaxy
The Majewski Lab ExomeAI Server supports detection of recurrent allelic imbalance in tumors using whole exome sequencing data, using ExomeAI, a free web-based application for detection of recurrent AI/LOH segments in tumor samples. Support is provided in the ExomeAI Manual, and via [email](mailto:Javad DOT Nadaf AT gmail DOT com). See Nadaf J, Majewski J, Fahiminiya S. (2014). ExomeAI: Detection of recurrent Allelic Imbalance in tumors using whole Exome sequencing data. Bioinformatics. 2014 Oct 8.
The Majewski Lab ExomeAI Server is supported by the McGill University and Génome Québec Innovation Centre
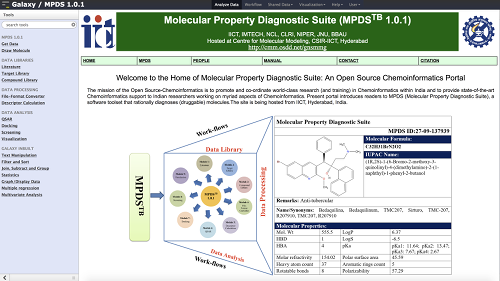
OSDD Molecular Property Diagnostic Suite (MPDS)
The OSDD Molecular Property Diagnostic Suite (MPDS) Galaxy server is an OSDD Chemoinformatics Portal. MPDS exposes a software toolset that rationally diagnoses (druggable) molecules. MPDS 1.0 consists of six modules covering informatics (DataBases, File format conversion), structure and analogue based drug design approaches (Property calculation, QSAR, Docking). Support is available.
MPDS is developed under the broad initiative of OSDD (Open Source Drug Discovery) of CSIR (Council of Scientific and Industrial Research, Govt. of India). The site is being hosted from IICT, Hyderabad, India.
Galaxy Community Hubs

Share your experience now
There were no new Log Board or Deployment Catalog entries in August! Eek! Please don't let this happen again!
The Community Log Board and Deployment Catalog Galaxy community hubs were launched last your. If you have a Galaxy deployment, or experience you want to share then please publish them this month.
New Releases
BioBlend v0.5.2 was released in October. BioBlend is a python library for interacting with CloudMan and the Galaxy API.
New versions of Galaxy, CloudMan, and blend4j were all released in August.
Look for a new Galaxy distribution in November.
ToolShed Contributions
Galaxy Project ToolShed Repos
Here are new contributions for the past two months.
In no particular order:
Tools
-
From crs4:
- seal_galaxy: Galaxy wrappers for Seal
-
From arkarachai-fungtammasan:
- microsatellite_ngs: Pipeline to profile and genotype microsatellites from short read data. This repository contains these sets of tools: 1 create microsatellite length profile, 2 correct for sequencing errors and report genotype, 3 estimate minimum sequencing read depth, 4 convert informative read depth to locus specific/genome wide sequencing depth.
-
From peterjc:
- mummer: v0.0.1, essentially a preview (previously only on the TestToolShed). A simple wrapper allowing MUMmer to be used to draw dotplots from within Galaxy using mummer, mucmer, or promer with mummerplot. No tests yet, no gnuplot or ps2pdf dependency yet.
-
From devteam:
- picard_plus: Picard wrappers for version 122 and up. New set of Picard wrappers that do not rely on external scripts and deal with all aspects of picard management and UI via tool XML.
-
From saket-choudhary:
- fathmm_web: Calls FATHMM webservice at http://fathmm.biocompute.org.uk
- mutationassessor_web: Call Mutation Assessor webservice
- replace_delimiters: Allows replacing any delimiter in the input to any other delimiter. This tool is similar to Galaxy's default 'Convert delimiter' tool, but allows conversion from any given type(comma, dash, pipe etc)
- inchlib_clust: a python script that performs data clustering and prepares input data for InCHlib. inchlib_clust can be used both from command line or from Python code. Data for clustering are supplied to inchlib_clust as a csv file.
- vep_rest: Variant Effect Predictor Webservice Package to interact with the GRCh37 (ONLY!). Variant Effect Predictor webservice at http://grch37.rest.ensembl.org
- chasm_webservice: Calls CHASM webserice at www.cravat.us
- polyphen2_web: Calls Polyphen2 webservice at http://genetics.bwh.harvard.edu/pph2/
- merge_columns_with_delimiter: Modified merge_columns to allow merging columns separated by a delimiter. This tool allows merging columns separated by a delimiter (two or multiple columns). It is similar to the Galaxy's default too; 'Merge Columns' but also allows merging them separated by a specified delimiter.
-
From galaxyp:
- pepxml_to_xls: Convert PepXML to Tabular
- protxml_to_xls: Convert ProtXML to Tabular
- blastxml_to_tabular_selectable: Converts blast xml file to a tabular with options for unmatched queries, and number of hits to convert. The unmatched queries can be useful for finding novel peptides.
- blast_plus_remote_blastp: NCBI BLAST+ remote blastp NCBI BLAST+ blastp with additional optional arguments.
Workflows
-
From bgruening:
- chemicaltoolbox_library_hole_filling_workflow: Uploaded Given one library, it extends all molecules by similar molecules of an other library and thus fill gaps in an automatic manner. Given one library, it extends all molecules by similar molecules of an other library and thus fill gaps in an automatic manner. This workflow is part of case study demonstrating the capability of the chemicaltoolbox. For further information please have a look at the chemicaltoolbox: https://github.com/bgruening/galaxytools/tree/master/chemicaltoolbox
Packages / Tool Dependency Definitions
-
From takadonet:
- package_tbl2asn_23_7: Contains a tool dependency definition that downloads the binary version 23.7 of tbl2asn. tbl2asn is an automated bulk submission program.
- package_minced_0_1_6: Contains a tool dependency definition that downloads version 0.1.6 of minced, a CRISPR finder. MinCED is a program to find Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) in full genomes or environmental datasets such as metagenomes, in which sequence size can be anywhere from 100 to 800 bp. MinCED runs from the command-line and was derived from CRT (http://www.room220.com/crt/): Charles Bland ''et al.'', CRISPR Recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats, BMC Bioinformatics 8, no. 1 (2007): 209.
- package_barrnap_0_5: Contains a tool dependency definition that downloads and compiles version 0.4 of the barrnap. Barrnap predicts the location of 5S, 16S and 23S ribosomal RNA genes in Bacterial genome sequ It takes FASTA DNA sequence as input, and write GFF3 as output. https://github.com/Victorian-Bioinformatics-Consortium/barrnap
-
From iuc:
- package_numpy_1_9: Contains a tool dependency definition that downloads and compiles version 1.9 of the the python numpy package. NumPy is the fundamental package for scientific computing with Python.
- package_blast_plus_2_2_30: first version, based on BLAST+ 2.2.29 definition. NCBI BLAST+ 2.2.30 (binaries only) This Tool Shed package is intended to be used as a dependency of the Galaxy wrappers for NCBI BLAST+ and any other tools which call the BLAST+ binaries internally.
- package_matplotlib_1_4: Contains a tool dependency definition that downloads and compiles version 1.4.x of the the python matplotlib package. matplotlib is a python 2D plotting library which produces publication quality figures. This is the version 1.2.x of matplotlib. www.matplotlib.org/
- package_networkx_1_9: Contains a tool dependency definition that downloads and compiles version 1.9.x of the python library networkx. NetworkX is a Python language software package for the creation, manipulation, and study of the structure, dynamics, and functions of complex networks. http://networkx.github.io/
- package_scipy_0_14: Contains a tool dependency definition that downloads and compiles version 0.14 of the the scipy python library. SciPy is open-source software for mathematics, science, and engineering. The SciPy library is built to work with NumPy arrays, and provides many user-friendly and efficient numerical routines such as routines for numerical integration and optimization. http://www.scipy.org/
- package_dill_0_2: Contains a tool dependency definition that downloads and compiles version 1.9.x of the python library dill. Dill extends python's 'pickle' module for serializing and de-serializing python objects to the majority of the built-in python types. Serialization is the process of converting an object to a byte stream, and the inverse of which is converting a byte stream back to on python object hierarchy. http://trac.mystic.cacr.caltech.edu/project/pathos/wiki/dill
- package_scikit_learn_0_15: Contains a tool dependency definition that downloads and compiles version 0.15.x of the the scikit-learn package. Easy-to-use and general-purpose machine learning in Python. Scikit-learn integrates machine learning algorithms in the tightly-knit scientific Python world, building upon numpy, scipy, and matplotlib. As a machine-learning module, it provides versatile tools for data mining and analysis in any field of science and engineering. It strives to be simple and efficient, accessible to everybody, and reusable in various contexts. http://scikit-learn.org/
-
From saket-choudhary:
- package_xlrd_0_9_3: Tool dependency definition of python-xlrd
- package_scikit_learn_0_15: Tool dependency package for scikit-learn-0.15
- package_fastcluster_1_1_13: Tool dependency definition of python-fastcluster
- package_blas_3_5_0: Tool dependency package for blas
- package_pyvcf_0_6_7: Tool dependedency definition for PyVCF
-
From lparsons:
- package_cutadapt_1_6: Initial version Contains a tool dependency definition that downloads and compiles cutadapt version 1.6 trim adapters from high-throughput sequencing reads
-
From devteam:
- package_picard_122: Picard 1.122 package definition This picard package dependency is retrieved directly from https://github.com/broadinstitute/picard/releases
Workflows
-
From bgruening:
- chemicaltoolbox_library_hole_filling_workflow: Uploaded Given one library, it extends all molecules by similar molecules of an other library and thus fill gaps in an automatic manner. Given one library, it extends all molecules by similar molecules of an other library and thus fill gaps in an automatic manner. This workflow is part of case study demonstrating the capability of the chemicaltoolbox. For further information please have a look at the chemicaltoolbox: https://github.com/bgruening/galaxytools/tree/master/chemicaltoolbox
Select Updates
-
From lparsons:
- cutadapt: Updated to version 1.6
- htseq_count: Deleted accidentally added file
-
From saskia-hiltemann:
- ireport: fixed dependencies and added MarkDown support
-
From devteam:
- quality_filter: tool definition that does not fail on stderr output.
-
From crs4:
- sspace: Update Orione citation. Update dependency to SSPACE Basic v2.1 . Add
. - prokka: Use
because prokka writes some warnings on stderr. Update Orione citation. Update Prokka citation. Support Prokka 1.10. Upgrade dependencies to package_minced_0_1_6, package_barrnap_0_5 and package_tbl2asn_23_7. Added --proteins option. Add .
- sspace: Update Orione citation. Update dependency to SSPACE Basic v2.1 . Add
-
From devteam:
- package_galaxy_ops_1_0_0: tool dependency definition that uses pip to install gops.
-
From miller-lab:
- package_quicktree_1_1: tool_dependencies.xml with a URL that works for urllib.
Other News
- Have you ever wanted to improve a Galaxy tool? Large number of tools are now awaiting your pull requests at GitHub.
- On the awesomeness of the BOSC/OpenBio Codefest 2014
- Björn Grüning's Galaxy Docker image now bundles FTP & SLURM and allows targeting external Galaxy directories.
- An IPython notebook demonstrating an interactive cummeRbund analysis embedded in Galaxy.
- GUGGO tutorial: how to activate Docker functionality in Galaxy; create a Docker image and install Stacks on that image; and integrate a Stacks Galaxy tool using that image.